Data filtering using R Studio
When you conduct statistical analysis, you might want to include/exclude some variables. For example, here is one data.
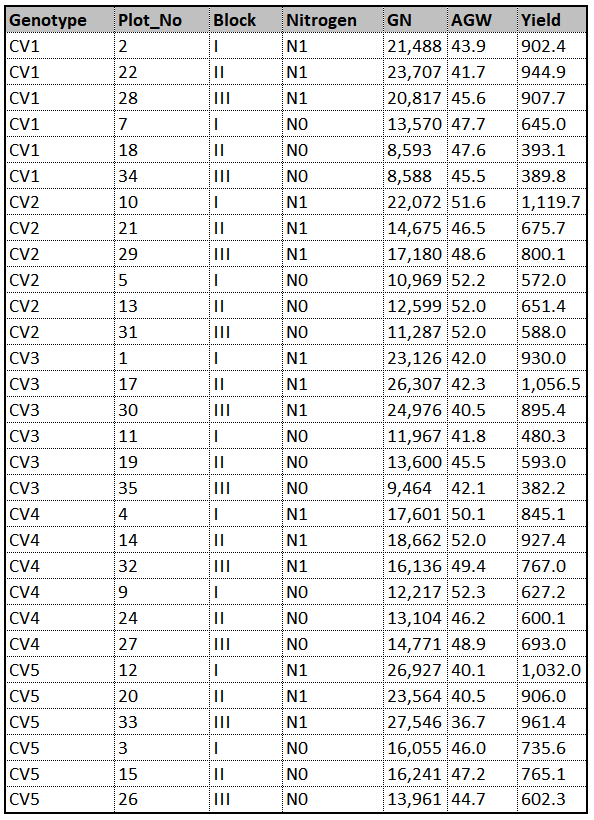
This is data about how yield, grain number (GN) and averge grain weight (AGW) are different according to two different fertilizers (N0, N1) in five genotypes (CV1 – CV5). That is, there will be 10 treatments [Genotype (5) x Nitrogen (2) =10]. Replicates are 10 as blocks, and therefore experimental unit will be 30 [10 treatments x 3 blocks = 30].
What if we want to analyze in only N1 condition? or only about CV1? I’ll introdce how to filter data in R studio?
Let’s upload the data above.
# to uplopad data
library (readr)
github="https://raw.githubusercontent.com/agronomy4future/raw_data_practice/main/yield%20component_nitrogen.csv"
dataA=data.frame(read_csv(url(github),show_col_types=FALSE))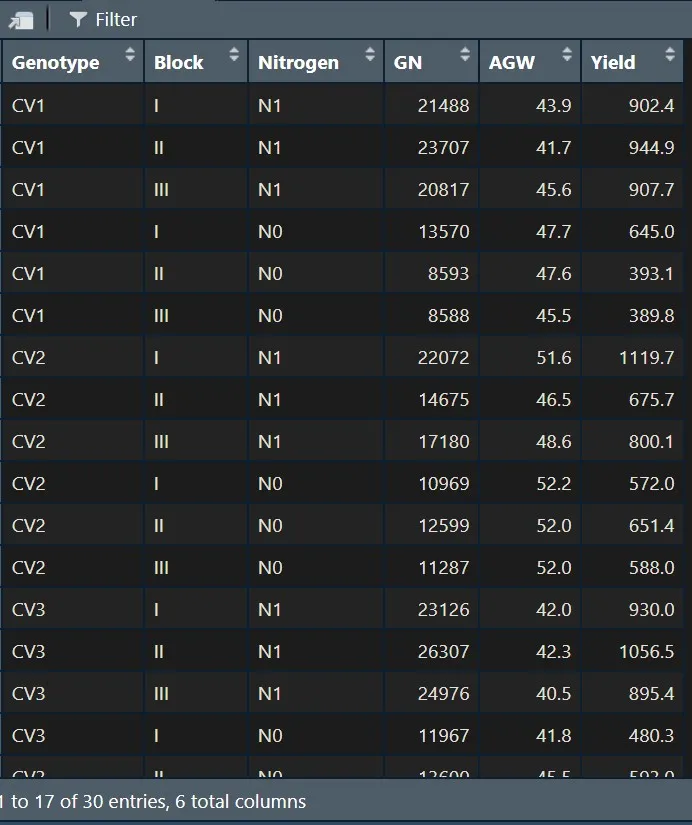
1. subset
I’d like to filter one variable. For example, there are two ways to select N1.
N1= subset (dataA, Nitrogen=="N1")
N1= subset (dataA, Nitrogen!="N1")
print(N1)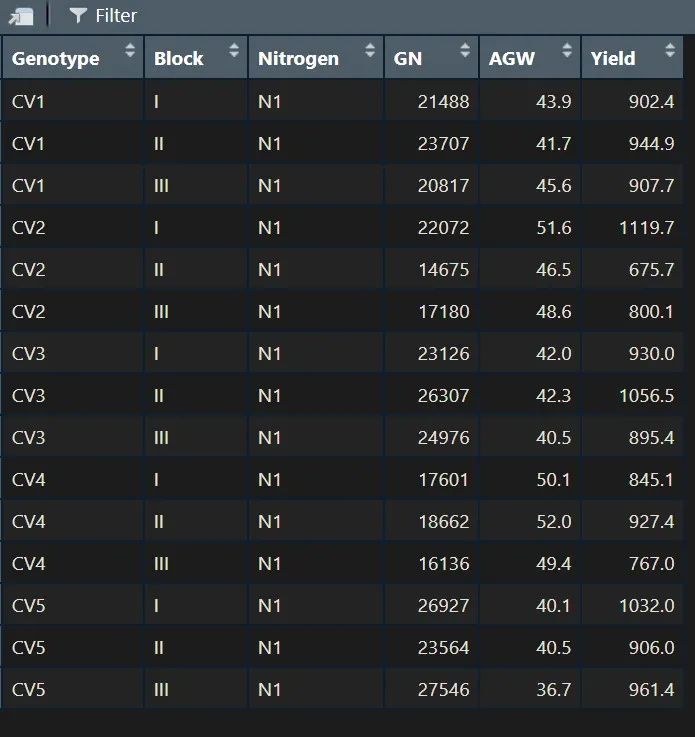
How about selecting several variables? For example, I want to select CV1 and N1.
CV1_N1= subset (dataA, Genotype=="CV1" & Nitrogen=="N1")
CV1_N1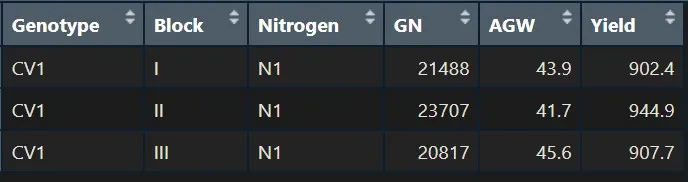
How about selecting one variable and excluding another variable?
CV1X_N1= subset (dataA, Genotype!="CV1" & Nitrogen=="N1")
CV1X_N1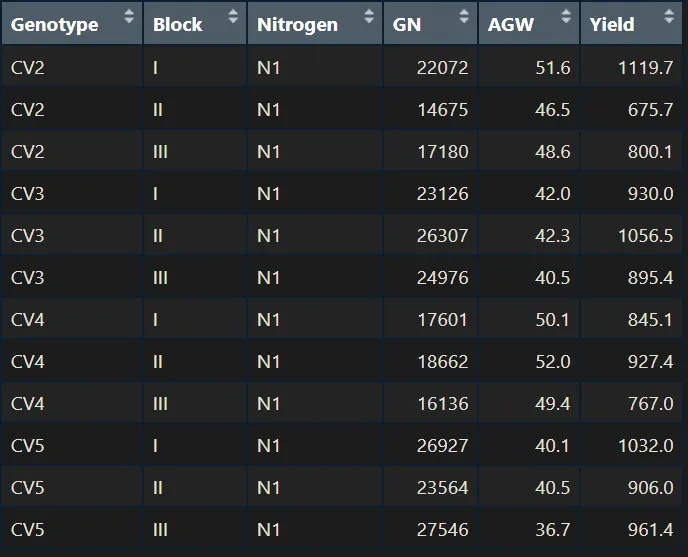
How about selecting two variables within the same factor? For example, I want to select both CV1 and CV3. So I used the below code.
CV1_CV3= subset (dataA, Genotype=="CV1" & Genotype=="CV3")
CV1_CV3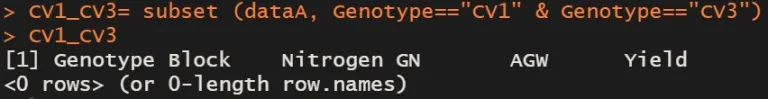
But I can’t select any variables. This is because if I select CV1, and now only CV1 exists. In this condition, if I select CV3 which does not exist, no variables are selected.
We can solve this problem using |
CV1_CV3= subset (dataA,Genotype=="CV1" | Genotype=="CV3")
CV1_CV3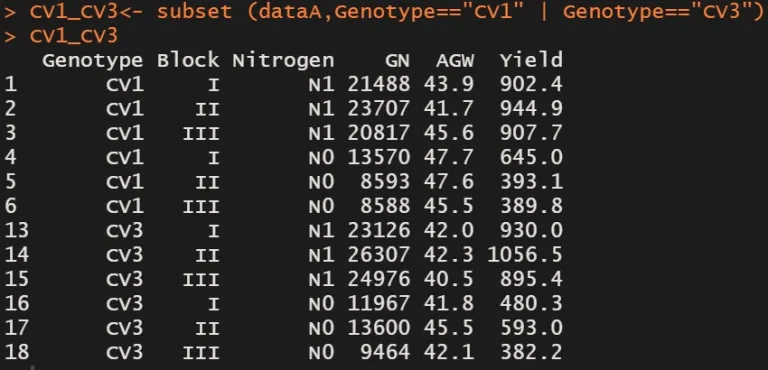
Or below code is possible.
CV1_CV3= subset (dataA, Genotype!="CV2" & Genotype!="CV4" & Genotype!="CV5" ) How about selecting two variables within the same factor, and another variable? For example, I want to select both CV1 and CV3, and then select N1.
CV1_CV3_N1= subset (dataA, c(Genotype=="CV1" | Genotype=="CV3") & Nitrogen=="N1")
CV1_CV3_N1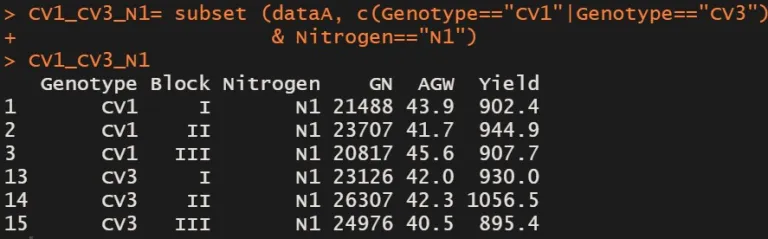
There are no specific answers. Simply we can use below code.
N1= subset (dataA, Nitrogen=="N1")
CV1_CV3_N1= subset(N1, Genotype=="CV1" | Genotype=="CV3")
CV1_CV3_N1First, we can select N1, and in N1, we can selecte both CV1 and CV3. We can shorten the code like below.
CV1_CV3_N1= subset(subset (dataA, Nitrogen=="N1"), Genotype=="CV1" | Genotype=="CV3")In summary, these three codes are the same code to select CV1, CV3, N1.
#1
CV1_CV3_N1= subset (dataA, c(Genotype=="CV1" | Genotype=="CV3") & Nitrogen=="N1")
#2
N1<- subset (dataA, Nitrogen=="N1")
CV1_CV3_N1= subset(N1, Genotype=="CV1" | Genotype=="CV3")
#3
CV1_CV3_N1= subset(subset(dataA, Nitrogen=="N1"), Genotype=="CV1" | Genotype=="CV3")2. dplyr
Now, let’s use dplyr package.
install.packages ("dplyr")
library (dplyr)Now, I’d like to select N1
#1
dataB= dataA %>% filter (Nitrogen=="N1")
#2
dataB= dataA %>% filter (Nitrogen!="N0")
print (dataB)
How about selecting CV1 and N1? It’s similar with subset()
#subset()
CV1_N1= subset (dataA, Genotype=="CV1" & Nitrogen=="N1")
#dplyr
CV1_N1= dataA %>% filter (Genotype=="CV1" & Nitrogen!="N0")How about selecting both CV1 and CV3?
#subset()
CV1_CV3= subset (dataA, Genotype=="CV1" | Genotype=="CV3")
#dplyr
CV1_CV3= dataA %>% filter(Genotype=="CV1"| Genotype=="CV3")How about selecting both CV1 and CV3, and then N1?
#subset()
CV1_CV3_N1= subset (dataA, c(Genotype=="CV1" | Genotype=="CV3") & Nitrogen=="N1")
#dplyr
CV1_CV3_N1= dataA %>% filter (c(Genotype=="CV1" | Genotype=="CV3") & Nitrogen=="N1")Now, let’s export this data to Excel.
install.packages("writexl")
library(writexl)
write_xlsx(CV1_CV3_N1,"C://Users/Usuari/Desktop//CV1_3_N1.xlsx")