Python Data Preprocessing: Practice
Before diving into in-depth data analysis, a crucial step is data preprocessing. This essential process not only ensures better data quality but also significantly improves the efficiency of your analysis. In this guide, I will introduce a range of powerful Python methods for data preprocessing, equipping you with the tools to optimize your data for more accurate and insightful analysis.
I use Goolge Colab when using Python because it’s more user friendly. Please refer how to setup Google Colab in below post.
□ How to use Google Colab for Python (power tool to analyze data)?
First, let’s mount your Google Colab environment.
To do this, use the following code:
from google.colab import drive drive.mount('/content/drive')Next, let’s proceed to import the data. I have uploaded a dataset to my GitHub repository. By copying and pasting the code snippet below, you can automatically download the data.”
import pandas as pd
url="https://raw.githubusercontent.com/agronomy4future/raw_data_practice/main/python_practice.csv"
dataA=pd.read_csv(url)Next, let’s see how the data is organized. First, I’ll see the top 15 rows.
print(dataA.head(15))
genotype height spike_weight disease_spot heat_tolerance region
0 CV1 86.5 27.90 0 yes Mid_East
1 CV2 85.5 33.77 1 no Test_field
2 CV2 95.5 33.00 3 no Test_field
3 CV2 100.5 22.71 0 no North
4 CV2 99.5 28.88 0 no North
5 CV1 98.5 25.74 0 no Test_field
6 CV1 113.5 33.44 1 no Test_field
7 CV1 104.5 27.74 3 no North
8 CV2 104.5 29.83 2 no Mid_West
9 CV1 127.5 25.84 0 no North
10 CV2 92.5 26.22 0 no Mid_West
11 CV1 129.5 26.29 0 yes Test_field
12 CV2 90.5 34.40 0 no Mid_East
13 CV1 123.5 39.82 0 no Test_field
14 CV2 94.5 NaN 0 yes Test_field
yield
0 16861.42400
1 1702.05230
2 4425.96200
3 21960.97061
4 3843.35520
5 3733.12160
6 8217.08960
7 7258.00560
8 6382.91070
9 28899.63692
10 2697.82080
11 27785.22510
12 1803.34300
13 11067.21780
14 39588.25770 Additionally, let’s check the number of rows and columns in this data, identify any duplicated entries, and resolve them by keeping one and removing the others.”
# to check the number of rows and columns
print(dataA.shape)
> (1338, 7)
# to see how many duplicated data exist
print("duplicated data No.", len(dataA[dataA.duplicated()]))
> duplicated data No. 1
# to see what the duplicated data are
print(dataA[dataA.duplicated(keep=False)].sort_values(by=list(dataA.columns)).head())
genotype height spike_weight disease_spot heat_tolerance region
195 CV2 86.5 30.59 0 no North
581 CV2 86.5 30.59 0 no North
yield
195 1616.0631
581 1616.0631
# to keep one and removing the others.
dataA.drop_duplicates(inplace=True, keep="first", ignore_index=True)Now, I’ll examine the structure of this data.
print(dataA.info())
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1337 entries, 0 to 1336
Data columns (total 7 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 genotype 1337 non-null object
1 height 1337 non-null float64
2 spike_weight 1332 non-null float64
3 disease_spot 1337 non-null int64
4 heat_tolerance 1337 non-null object
5 region 1337 non-null object
6 yield 1337 non-null float64
dtypes: float64(3), int64(1), object(3)I’ll create a graph to visualize the number of categorical values.
import seaborn as sns
import matplotlib.pyplot as plt
custom_palette = ["darkred", "darkblue"]
for col in dataA.select_dtypes(include=['object', 'category']).columns:
fig = sns.catplot(x=col, kind="count", data=dataA, hue=None, palette=custom_palette)
fig.set_xticklabels(rotation=90)
plt.show() 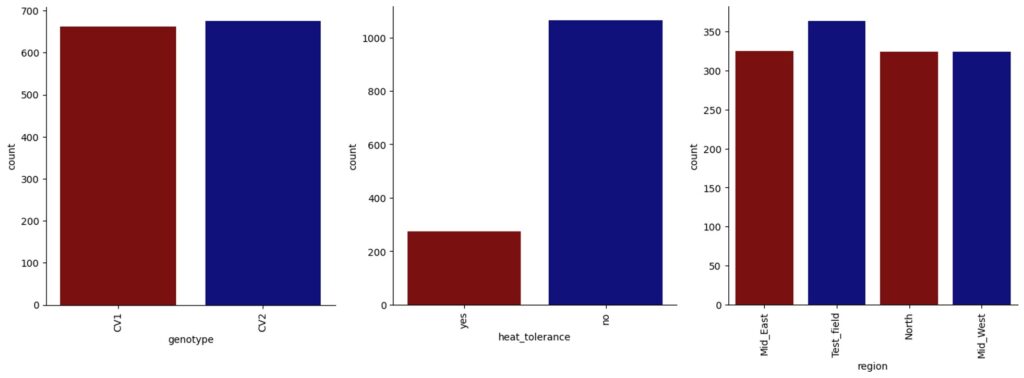
python_code/data_check_process.ipynb at main · agronomy4future/python_code (github.com)