A Guide to Normalizing Data for Different Treatments in R
I have data, as shown below, regarding iron contents in soil and the plant uptake of iron at different growth stages in winter wheat. I want to analyze the relationship between the iron content in the soil and the plant uptake of iron at different growth stages in winter wheat.
library(readr)
github="https://raw.githubusercontent.com/agronomy4future/raw_data_practice/main/wheat_grain_Fe_uptake.csv"
dataA= data.frame(read_csv(url(github), show_col_types= FALSE))
Location Season Genotype Reps Iron_ton_ha Stage Fe
1 East 2021 CV1 1 21.7127 Vegetative 0.44
2 East 2021 CV1 2 8.7340 Vegetative 0.30
3 East 2021 CV1 3 9.5003 Vegetative 0.31
4 East 2021 CV1 4 5.9481 Vegetative 0.37
5 East 2021 CV1 5 7.4608 Vegetative 0.30
6 East 2021 CV1 6 20.5326 Vegetative 0.33
7 East 2021 CV1 7 19.8532 Vegetative 0.29
8 East 2021 CV1 8 7.9718 Vegetative 0.35
9 East 2021 CV1 9 15.0087 Vegetative 0.38
10 East 2021 CV1 10 5.6608 Vegetative 0.40
.
.
.We can simply draw a regression graph. However, before doing that, we need to reshape the data. I’ll transpose the data from rows to columns based on the variables in the Stage column.
library(dplyr)
library(tidyr)
dataB= data.frame(dataA %>%
group_by(Location, Season, Genotype, Iron_ton_ha, Stage) %>%
spread(key=Stage, value = Fe))
dataB
Location Season Genotype Iron_ton_ha Maturity Reproductive Vegetative
1 East 2021 CV1 5.6608 0.14 0.31 0.40
2 East 2021 CV1 5.9481 0.18 0.30 0.37
3 East 2021 CV1 6.2757 0.08 0.29 0.38
4 East 2021 CV1 7.4608 0.12 0.34 0.30
5 East 2021 CV1 7.5574 0.15 0.28 0.36
6 East 2021 CV1 7.8064 0.15 0.29 0.34
7 East 2021 CV1 7.9718 0.17 0.31 0.35
8 East 2021 CV1 8.0175 0.15 0.30 0.45
9 East 2021 CV1 8.2875 0.14 0.31 0.36
10 East 2021 CV1 8.6052 0.11 0.32 0.38
.
.
.Now, I will analyze the relationship between the iron (Fe) content in the soil and the uptake of iron by plants at maturity.
library(ggplot2)
FIGA= ggplot(data=dataB, aes(x=Iron_ton_ha, y=Reproductive))+
geom_point(aes(fill=as.factor(Season), shape=as.factor(Season)),color="black", size=5) +
scale_fill_manual(values= c("grey15","grey35","grey55")) +
scale_shape_manual(values= c(21,22,24)) +
scale_x_continuous(breaks=seq(0,60,20),limits=c(0,60)) +
scale_y_continuous(breaks=seq(0,0.5,0.1),limits=c(0,0.5)) +
facet_wrap(~ Location) +
annotate("segment", x=20, xend=40, y=Inf, yend=Inf, color="black",lwd=1)+
labs(x="Fe (Mg/ha) in soil", y="Plant Fe uptake (%) at maturity") +
theme_classic(base_size=18, base_family="serif") +
theme(legend.position=c(0.90,0.17),
legend.title=element_blank(),
legend.key=element_rect(color="white", fill="white"),
legend.text=element_text(family="serif", face="plain", size=13, color="black"),
legend.background= element_rect(fill="white"),
strip.background=element_rect(color="white", linewidth=0.5, linetype="solid"),
axis.line = element_line(linewidth = 0.5, colour="black"))
FIGA + windows(width=9, height=5)
ggsave("C:/Users/dream/Desktop/R_OUTPUT/FIGA.jpg",
FIGA, width=9*2.54, height=5*2.54, units="cm", dpi=1000)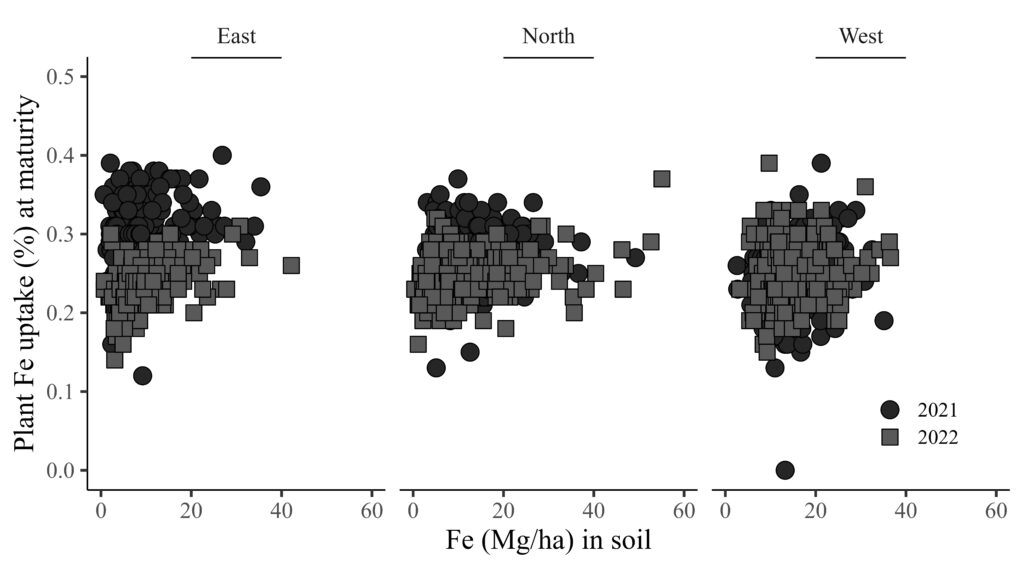
Now, I’ll normalize the data for each treatment combination (i.e., East, 2021, CV1) using the mutate() function. For easier normalization calculations, a vertically organized data format is preferable. Therefore, I’ll be using the dataA dataset.
dataC= data.frame(dataA %>%
group_by(Location, Season, Genotype) %>%
mutate(
Normalized_Fe_plant=(Fe-mean(Fe))/sd(Fe),
Normalized_Fe_soil=(Iron_ton_ha-mean(Iron_ton_ha))/sd(Iron_ton_ha)
))
dataC
Location Season Genotype Reps Iron_ton_ha Stage Fe Normalized_Fe_plant Normalized_Fe_soil
1 East 2021 CV1 1 21.7127 Vegetative 0.44 1.682394399 1.85849093
2 East 2021 CV1 2 8.7340 Vegetative 0.30 0.246204058 -0.48486395
3 East 2021 CV1 3 9.5003 Vegetative 0.31 0.348789083 -0.34650550
4 East 2021 CV1 4 5.9481 Vegetative 0.37 0.964299229 -0.98786906
5 East 2021 CV1 5 7.4608 Vegetative 0.30 0.246204058 -0.71474518Then, let’s reshape the data from row to column regarding the Stage.
library(dplyr)
library(tidyr)
dataD= data.frame(dataC %>%
group_by(Location, Season, Genotype, Normalized_Fe_soil, Stage) %>%
spread(key=Stage, value= Normalized_Fe_plant))
dataD
Location Season Genotype Normalized_Fe_soil Maturity Reproductive Vegetative
1 East 2021 CV1 -1.03974219 -1.39515633 0.348789083 1.272054302
2 East 2021 CV1 -0.98786906 -0.98481623 0.246204058 0.964299229
3 East 2021 CV1 -0.92871960 -2.01066648 0.143619034 1.066884253
4 East 2021 CV1 -0.71474518 -1.60032638 0.656544156 0.246204058
5 East 2021 CV1 -0.69730367 -1.29257131 0.041034010 0.861714204Let’s draw the graph again.
library(ggplot2)
FIGB= ggplot(data=dataD, aes(x=Normalized_Fe_soil, y=Maturity))+
geom_point(aes(fill=as.factor(Season), shape=as.factor(Season)),color="black", size=5) +
scale_fill_manual(values= c("grey15","grey35","grey55")) +
scale_shape_manual(values= c(21,22,24)) +
scale_x_continuous(breaks=seq(-5,5,2.5),limits=c(-5,5)) +
scale_y_continuous(breaks=seq(-5,5,2.5),limits=c(-5,5)) +
geom_vline(xintercept=0, linetype="dashed", color="black") +
geom_hline(yintercept=0, linetype="dashed", color= "black") +
facet_wrap(~ Location) +
annotate("segment", x=20, xend=40, y=Inf, yend=Inf, color="black",lwd=1)+
labs(x="Fe (Mg/ha) in soil", y="Plant Fe uptake (%) at maturity") +
theme_classic(base_size=18, base_family="serif") +
theme(legend.position=c(0.90,0.17),
legend.title=element_blank(),
legend.key=element_rect(color="white", fill="white"),
legend.text=element_text(family="serif", face="plain", size=13, color="black"),
legend.background= element_rect(fill="white"),
strip.background=element_rect(color="white", linewidth=0.5, linetype="solid"),
axis.line = element_line(linewidth = 0.5, colour="black"))
FIGB + windows(width=9, height=5)
ggsave("C:/Users/dream/Desktop/R_OUTPUT/FIGB.jpg",
FIGB, width=9*2.54, height=5*2.54, units="cm", dpi=1000)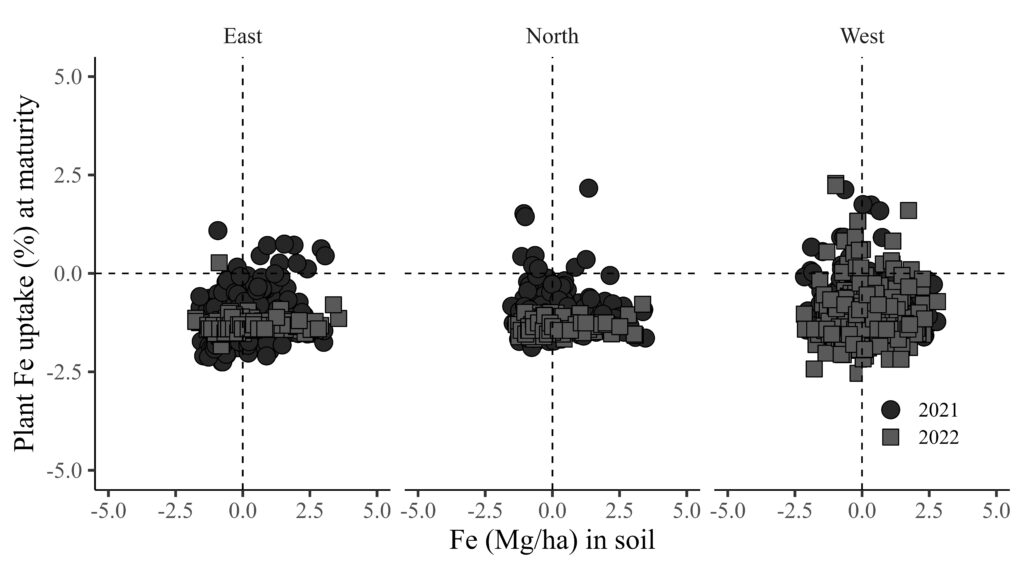
### full code
https://github.com/agronomy4future/r_code/blob/main/data_normalization
library(readr)
library(dplyr)
library(tidyr)
library(ggplot2)
#data upload
github="https://raw.githubusercontent.com/agronomy4future/raw_data_practice/main/wheat_Fe_uptake.csv"
dataA= data.frame(read_csv(url(github), show_col_types= FALSE))
# data normalization
dataB= data.frame(dataA %>%
group_by(Location, Season, Genotype) %>%
mutate(
Normalized_Fe_plant=(Fe-mean(Fe))/sd(Fe),
Normalized_Fe_soil=(Iron_ton_ha-mean(Iron_ton_ha))/sd(Iron_ton_ha)
))
# data reshape
dataC= data.frame(dataB %>%
group_by(Location, Season, Genotype, Normalized_Fe_soil, Stage) %>%
spread(key=Stage, value= Normalized_Fe_plant))
# graph
FIGB= ggplot(data=dataC, aes(x=Normalized_Fe_soil, y=Maturity))+
geom_point(aes(fill=as.factor(Season), shape=as.factor(Season)),color="black", size=5) +
scale_fill_manual(values= c("grey15","grey35","grey55")) +
scale_shape_manual(values= c(21,22,24)) +
scale_x_continuous(breaks=seq(-5,5,2.5),limits=c(-5,5)) +
scale_y_continuous(breaks=seq(-5,5,2.5),limits=c(-5,5)) +
geom_vline(xintercept=0, linetype="dashed", color="black") +
geom_hline(yintercept=0, linetype="dashed", color= "black") +
facet_wrap(~ Location) +
annotate("segment", x=20, xend=40, y=Inf, yend=Inf, color="black",lwd=1)+
labs(x="Fe (Mg/ha) in soil", y="Plant Fe uptake (%) at maturity") +
theme_classic(base_size=18, base_family="serif") +
theme(legend.position=c(0.90,0.17),
legend.title=element_blank(),
legend.key=element_rect(color="white", fill="white"),
legend.text=element_text(family="serif", face="plain", size=13, color="black"),
legend.background= element_rect(fill="white"),
strip.background=element_rect(color="white", linewidth=0.5, linetype="solid"),
axis.line = element_line(linewidth = 0.5, colour="black"))
FIGB + windows(width=9, height=5)Is R calculation correct?
I have just used the code and now I’m wondering whether the code I used correctly calculates normalization. Therefore, I will manually calculate normalization and compare the values.
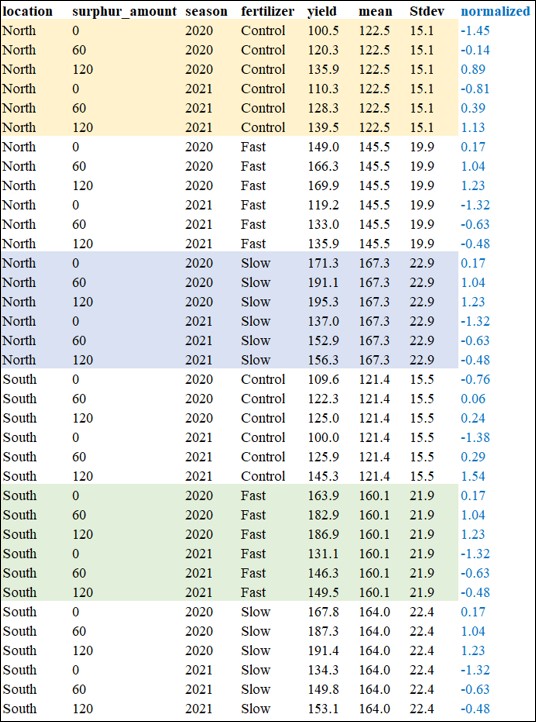
I pooled data from two seasons and normalized yield based on location and fertilizer. Now, I’ll check whether this normalized data matches what R calculated.
library(readr)
github="https://raw.githubusercontent.com/agronomy4future/raw_data_practice/main/normalization_practice.csv"
dataA= data.frame(read_csv(url(github), show_col_types= FALSE))
dataB= data.frame(dataA %>%
group_by(location, fertilizer) %>%
mutate(
Normalized_R=(normalized-mean(normalized))/sd(normalized),
))
dataB$Normalized_R=round(dataB$Normalized_R,digits=2)
dataB
location surphur_amount season fertilizer yield mean Stdev normalized Normalized_R
1 North 0 2020 Control 100.5 122.5 15.1 -1.45 -1.45
2 North 60 2020 Control 120.3 122.5 15.1 -0.14 -0.14
3 North 120 2020 Control 135.9 122.5 15.1 0.89 0.89
4 North 0 2021 Control 110.3 122.5 15.1 -0.81 -0.81
5 North 60 2021 Control 128.3 122.5 15.1 0.39 0.39
6 North 120 2021 Control 139.5 122.5 15.1 1.13 1.13
7 North 0 2020 Fast 149.0 145.5 19.9 0.17 0.17
8 North 60 2020 Fast 166.3 145.5 19.9 1.04 1.04
9 North 120 2020 Fast 169.9 145.5 19.9 1.23 1.23
10 North 0 2021 Fast 119.2 145.5 19.9 -1.32 -1.32
11 North 60 2021 Fast 133.0 145.5 19.9 -0.63 -0.63
12 North 120 2021 Fast 135.9 145.5 19.9 -0.48 -0.48
13 North 0 2020 Slow 171.3 167.3 22.9 0.17 0.17
14 North 60 2020 Slow 191.1 167.3 22.9 1.04 1.04
15 North 120 2020 Slow 195.3 167.3 22.9 1.23 1.23
16 North 0 2021 Slow 137.0 167.3 22.9 -1.32 -1.32
17 North 60 2021 Slow 152.9 167.3 22.9 -0.63 -0.63
18 North 120 2021 Slow 156.3 167.3 22.9 -0.48 -0.48
19 South 0 2020 Control 109.6 121.4 15.5 -0.76 -0.76
20 South 60 2020 Control 122.3 121.4 15.5 0.06 0.06
21 South 120 2020 Control 125.0 121.4 15.5 0.24 0.24
22 South 0 2021 Control 100.0 121.4 15.5 -1.38 -1.38
23 South 60 2021 Control 125.9 121.4 15.5 0.29 0.29
24 South 120 2021 Control 145.3 121.4 15.5 1.54 1.54
25 South 0 2020 Fast 163.9 160.1 21.9 0.17 0.17
26 South 60 2020 Fast 182.9 160.1 21.9 1.04 1.04
27 South 120 2020 Fast 186.9 160.1 21.9 1.23 1.23
28 South 0 2021 Fast 131.1 160.1 21.9 -1.32 -1.32
29 South 60 2021 Fast 146.3 160.1 21.9 -0.63 -0.63
30 South 120 2021 Fast 149.5 160.1 21.9 -0.48 -0.48
31 South 0 2020 Slow 167.8 164.0 22.4 0.17 0.17
32 South 60 2020 Slow 187.3 164.0 22.4 1.04 1.04
33 South 120 2020 Slow 191.4 164.0 22.4 1.23 1.23
34 South 0 2021 Slow 134.3 164.0 22.4 -1.32 -1.32
35 South 60 2021 Slow 149.8 164.0 22.4 -0.63 -0.63
36 South 120 2021 Slow 153.1 164.0 22.4 -0.48 -0.48My calculation matches the R calculation, so I can trust the code.
