Enhancing Visualizations: Manipulating Color and Shape in R with Two Variables
I have one dataset as below.
library(readr)
github="https://raw.githubusercontent.com/agronomy4future/raw_data_practice/main/genotype_transgenic_line_with_fertilizer.csv"
dataA= data.frame(read_csv(url(github), show_col_types= FALSE))
dataA
genotype plot resistance fertilizer GN AGW
1 cv1 1 no 150 1310 33.15
2 cv1 1 no 0 1168 25.07
3 cv1 1 no 200 570 36.04
4 cv2 1 no 150 1120 53.05
5 cv2 1 no 0 1230 26.71
6 cv2 1 no 200 795 37.75
7 cv3 1 yes 150 2013 27.51
8 cv3 1 yes 0 2183 24.61
9 cv3 1 yes 200 1034 36.53
10 cv4 1 no 150 2451 31.58
.
.
.Now, I’ll create a regression graph between grain number (GN) and average grain weight (AGW).
library(ggplot2)
FIG1=ggplot(data=dataA, aes(x=GN, y=AGW))+
geom_point(aes(fill=genotype, shape=genotype), color= "black", size= 5) +
geom_smooth(method='lm', linetype=1, se=FALSE, color="red", formula=y~x, size=0.5) +
scale_fill_manual(values=c("blue","red","orange","purple","green","black"))+
scale_shape_manual(values=rep(c(21),6))+
scale_x_continuous(breaks=seq(0,5000,1000), limits = c(0,5000)) +
scale_y_continuous(breaks=seq(0,80,20), limits = c(0,80)) +
labs(x="Grain number per panicle", y="Average grain weight (mg) per panicle") +
theme_classic(base_size=18, base_family="serif")+
theme(legend.position=c(0.8,0.8),
legend.title=element_blank(),
legend.key=element_rect(color="white", fill="white"),
legend.text=element_text(family="serif", face="plain",
size=13, color= "Black"),
legend.background=element_rect(fill="white"),
axis.line=element_line(linewidth=0.5, colour="black"),
strip.background=element_rect(color="white",
linewidth=0.5,linetype="solid"))
FIG1+windows(width=5.5, height=5)
ggsave("C:/Users/Desktop/R_OUTPUT/FIG1.jpg",
FIG1, width=5.5*2.54, height=5*2.54, units="cm", dpi=1000)
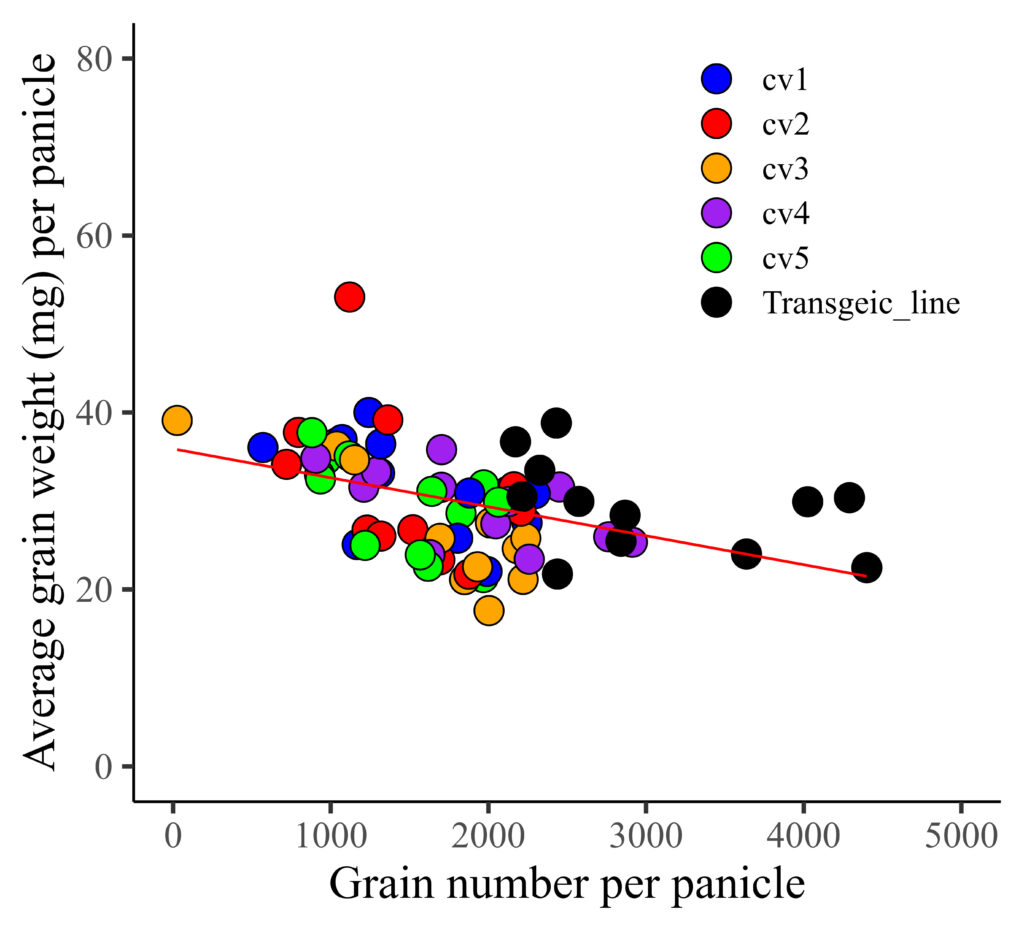
I distinguished genotypes with different colors, and now I want to differentiate resistance (yes and no) using distinct shapes. Therefore, I’ll be changing the shape representation from genotype to resistance.
library(ggplot2)
FIG2=ggplot(data=dataA, aes(x=GN, y=AGW))+
geom_point(aes(fill=genotype, shape=resistance), color= "black", size= 5) +
geom_smooth(method='lm', linetype=1, se=FALSE, color="red", formula=y~x, size=0.5) +
scale_fill_manual(values=c("blue","red","orange","purple","green","black"))+
scale_shape_manual(values=c(21,22)) +
scale_x_continuous(breaks=seq(0,5000,1000), limits = c(0,5000)) +
scale_y_continuous(breaks=seq(0,80,20), limits = c(0,80)) +
labs(x="Grain number per panicle", y="Average grain weight (mg) per panicle") +
theme_classic(base_size=18, base_family="serif")+
theme(legend.position=c(0.8,0.8),
legend.title=element_blank(),
legend.key=element_rect(color="white", fill="white"),
legend.text=element_text(family="serif", face="plain",
size=13, color= "Black"),
legend.background=element_rect(fill="white"),
axis.line=element_line(linewidth=0.5, colour="black"),
strip.background=element_rect(color="white",
linewidth=0.5,linetype="solid"))
FIG2+windows(width=5.5, height=5)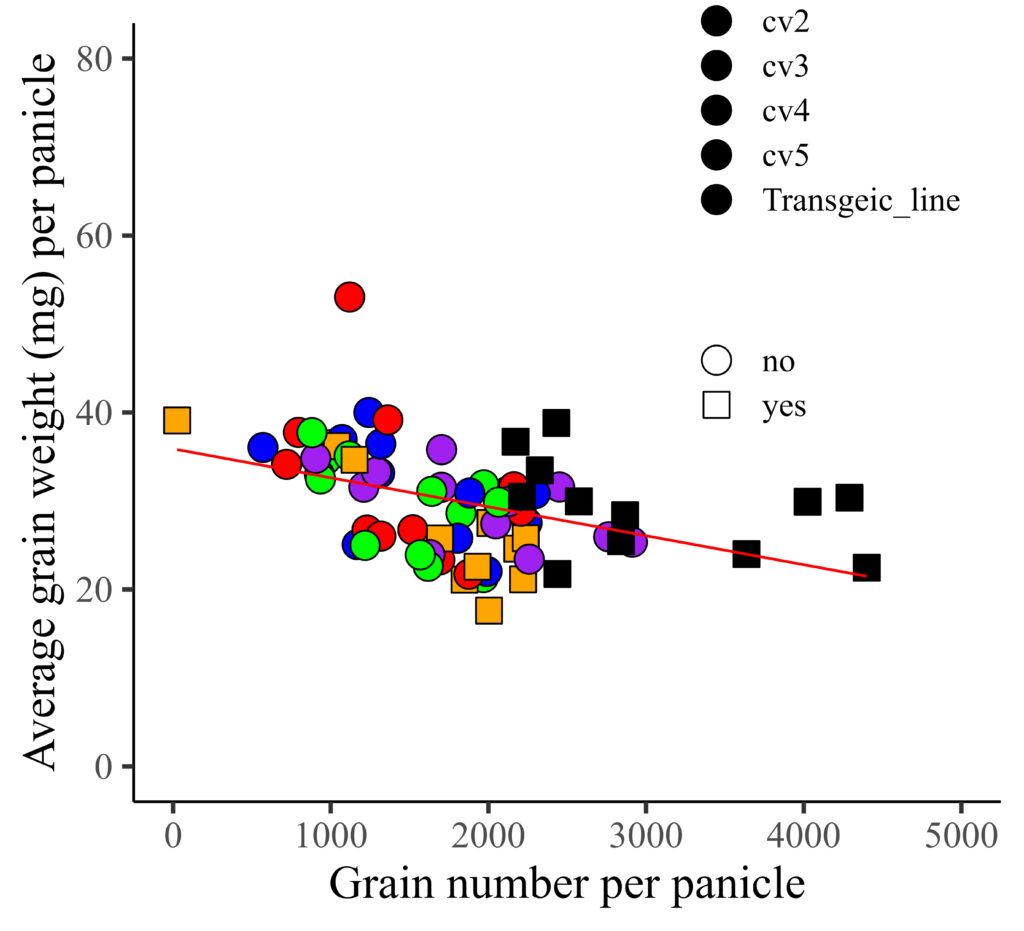
However, the color is not currently applied to the legend. I aim to apply the provided color to the legend, and additionally, assign colors to represent different levels of resistance.
Here is the solution!!
library(ggplot2)
FIG3=ggplot(data=dataA, aes(x=GN, y=AGW))+
geom_point(aes(fill=genotype, shape=resistance), color= "black", size= 5) +
geom_smooth(method='lm', linetype=1, se=FALSE, color="red", formula=y~x, size=0.5) +
scale_fill_manual(values=c("tomato1","tomato3","tomato4","blue","darkblue","black"),
guide=guide_legend(override.aes=list(shape=21))) +
scale_shape_manual(values=c(24,22),
guide=guide_legend(override.aes=list(colour="black"))) +
scale_x_continuous(breaks=seq(0,5000,1000), limits = c(0,5000)) +
scale_y_continuous(breaks=seq(0,80,20), limits = c(0,80)) +
labs(x="Grain number per panicle", y="Average grain weight (mg) per panicle") +
theme_classic(base_size=18, base_family="serif")+
theme(legend.position=c(0.8,0.8),
legend.title=element_blank(),
legend.key=element_rect(color="white", fill="white"),
legend.text=element_text(family="serif", face="plain",
size=13, color= "Black"),
legend.background=element_rect(fill="white"),
axis.line=element_line(linewidth=0.5, colour="black"),
strip.background=element_rect(color="white",
linewidth=0.5,linetype="solid"))
FIG3+windows(width=5.5, height=5)
ggsave("C:/Users/dream/Desktop/R_OUTPUT/FIG3.jpg",
FIG3, width=5.5*2.54, height=5*2.54, units="cm", dpi=1000)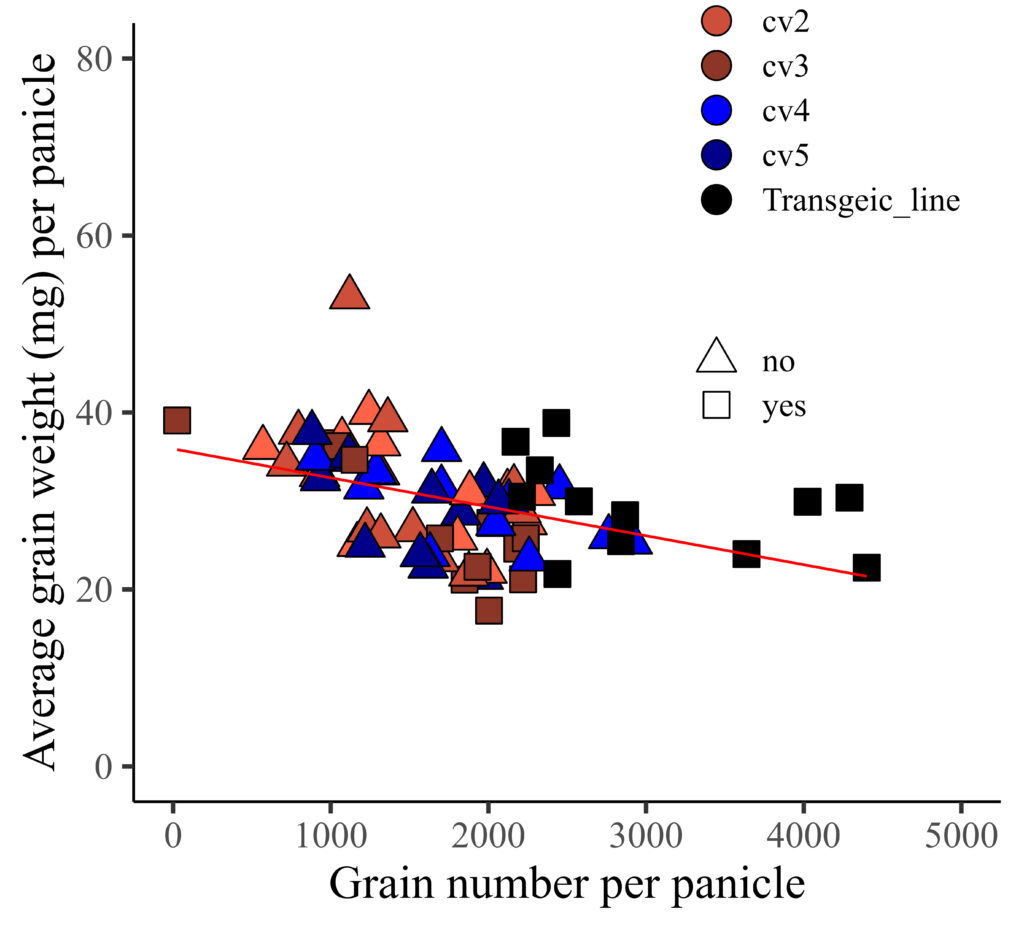
If you want to display two legend horizontally, you can add legend.box="horizontal"
library(ggplot2)
FIG4=ggplot(data=dataA, aes(x=GN, y=AGW))+
geom_point(aes(fill=genotype, shape=resistance), color= "black", size= 5) +
geom_smooth(method='lm', linetype=1, se=FALSE, color="red", formula=y~x, size=0.5) +
scale_fill_manual(values=c("tomato1","tomato3","tomato4","blue","darkblue","black"),
guide=guide_legend(override.aes=list(shape=21))) +
scale_shape_manual(values=c(24,22),
guide=guide_legend(override.aes=list(colour="black"))) +
scale_x_continuous(breaks=seq(0,5000,1000), limits = c(0,5000)) +
scale_y_continuous(breaks=seq(0,80,20), limits = c(0,80)) +
labs(x="Grain number per panicle", y="Average grain weight (mg) per panicle") +
theme_classic(base_size=18, base_family="serif")+
theme(legend.position=c(0.65,0.8),
legend.box= "horizontal",
legend.title=element_blank(),
legend.key=element_rect(color="white", fill="white"),
legend.text=element_text(family="serif", face="plain",
size=13, color= "Black"),
legend.background=element_rect(fill="white"),
axis.line=element_line(linewidth=0.5, colour="black"),
strip.background=element_rect(color="white",
linewidth=0.5, linetype="solid"))
FIG4+windows(width=5.5, height=5)
ggsave("C:/Users/dream/Desktop/R_OUTPUT/FIG4.jpg",
FIG4, width=5.5*2.54, height=5*2.54, units="cm", dpi=1000)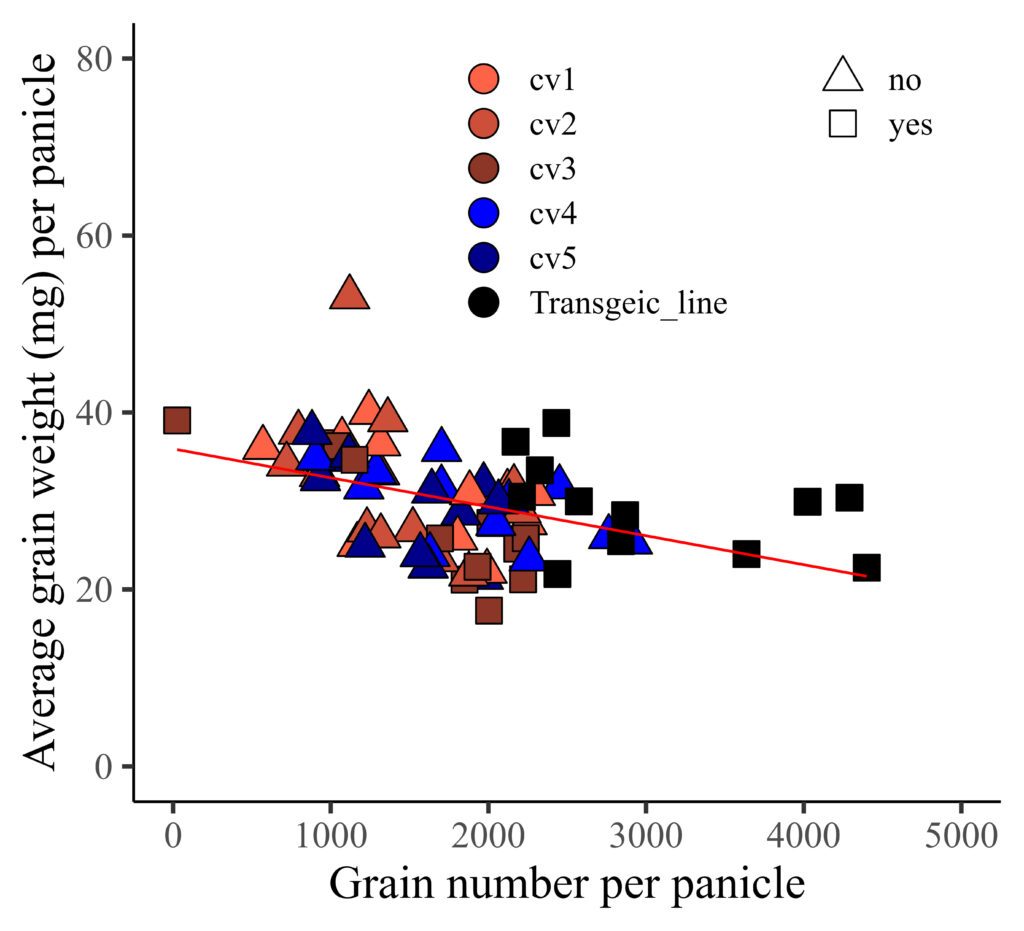
full code; https://github.com/agronomy4future/raw_data_practice/blob/main/different_color_shape_legend
library(readr)
library(ggplot2)
github="https://raw.githubusercontent.com/agronomy4future/raw_data_practice/main/genotype_transgenic_line_with_fertilizer.csv"
dataA= data.frame(read_csv(url(github), show_col_types= FALSE))
FIG4=ggplot(data=dataA, aes(x=GN, y=AGW))+
geom_point(aes(fill=genotype, shape=resistance), color= "black", size= 5) +
geom_smooth(method='lm', linetype=1, se=FALSE, color="red", formula=y~x, size=0.5) +
scale_fill_manual (values=c("tomato1","tomato3","tomato4","blue","darkblue","black"), guide=guide_legend(override.aes=list(shape=21))) +
scale_shape_manual(values=c(24,22), guide=guide_legend(override.aes=list(colour="black"))) +
scale_x_continuous(breaks=seq(0,5000,1000), limits = c(0,5000)) +
scale_y_continuous(breaks=seq(0,80,20), limits = c(0,80)) +
labs(x="Grain number per panicle", y="Average grain weight (mg) per panicle") +
theme_classic(base_size=18, base_family="serif")+
theme(legend.position=c(0.65,0.8),
legend.box= "horizontal",
legend.title=element_blank(),
legend.key=element_rect(color="white", fill="white"),
legend.text=element_text(family="serif", face="plain",
size=13, color= "Black"),
legend.background=element_rect(fill="white"),
axis.line=element_line(linewidth=0.5, colour="black"),
strip.background=element_rect(color="white",
linewidth=0.5, linetype="solid"))
FIG4+windows(width=5.5, height=5)
ggsave("C:/Users/dream/Desktop/R_OUTPUT/FIG4.jpg",
FIG4, width=5.5*2.54, height=5*2.54, units="cm", dpi=1000)
© 2022 – 2023 https://agronomy4future.com