Efficient Data Management: Variable Filtering in SAS Studio
Today, I’ll introduce how to filter variables after uploading data to SAS Studio. First, let’s upload data to SAS Studio.
library(readr)
github="https://raw.githubusercontent.com/agronomy4future/raw_data_practice/main/sorghum_grain.csv"
dataA= data.frame(read_csv(url(github), show_col_types=FALSE))
dataA
Location Crop Plot Block Treatment Treatment_modified Total_DW Grain_yield AGW GN
1 Location_A Sorghum 15 I Control Control 852.3 4303.6 28.1 31627
2 Location_A Sorghum 17 II Control Control 1295.7 6058.4 26.3 40157
3 Location_A Sorghum 19 III Control Control 1488.4 7438.7 20.8 45417
4 Location_A Sorghum 22 IV Control Control 925.6 5001.3 20.8 20770
5 Location_A Sorghum 1 I TR1 TR1_2 807.4 3285.2 30.7 14488
6 Location_A Sorghum 4 II TR1 TR1_2 907.4 3896.1 28.5 20323
7 Location_A Sorghum 9 III TR1 TR1_2 778.6 3207.3 28.8 15636
8 Location_A Sorghum 10 IV TR1 TR1_2 895.2 4418.2 28.6 20807
9 Location_A Sorghum 1 I TR2 TR1_2 748.9 3130.9 49.1 11556
10 Location_A Sorghum 4 II TR2 TR1_2 1032.4 4809.1 24.8 16821
11 Location_A Sorghum 9 III TR2 TR1_2 1388.2 6409.9 26.7 32546
12 Location_A Sorghum 10 IV TR2 TR1_2 1215.1 5183.3 29.3 20720
13 Location_A Sorghum 1 I TR3 TR3 668.7 3008.1 25.5 10390
14 Location_A Sorghum 4 II TR3 TR3 1564.1 7160.5 23.9 42890
15 Location_A Sorghum 9 III TR3 TR3 1283.4 6284.1 23.9 23713
16 Location_A Sorghum 10 IV TR3 TR3 1612.0 7445.1 20.9 34173
17 Location_A Sorghum 1 I TR4 TR4_5 1010.5 4340.3 29.2 23492
18 Location_A Sorghum 4 II TR4 TR4_5 757.4 2865.3 29.2 17564
19 Location_A Sorghum 9 III TR4 TR4_5 1087.1 5106.0 25.4 30184
20 Location_A Sorghum 10 IV TR4 TR4_5 1167.7 5383.6 27.7 23543
21 Location_A Sorghum 1 I TR5 TR4_5 783.3 2765.0 26.5 22890
22 Location_A Sorghum 4 II TR5 TR4_5 1002.5 3965.2 28.8 20570
23 Location_A Sorghum 9 III TR5 TR4_5 733.4 3445.1 29.5 15980
24 Location_A Sorghum 10 IV TR5 TR4_5 1067.9 5000.5 27.7 28911I’ll summarize data based on Treatment_modified.
library(dplyr)
summary=data.frame(dataA %>%
group_by(Location,Block, Treatment_modified) %>%
dplyr::summarize(across(c(Grain_yield,GN,AGW),
.fns= list(Mean=~mean(., na.rm=TRUE)))))
summary
Location Block Treatment_modified Grain_yield_Mean GN_Mean AGW_Mean
1 Location_A I Control 4303.60 31627.0 28.10
2 Location_A I TR1_2 3208.05 13022.0 39.90
3 Location_A I TR3 3008.10 10390.0 25.50
4 Location_A I TR4_5 3552.65 23191.0 27.85
5 Location_A II Control 6058.40 40157.0 26.30
6 Location_A II TR1_2 4352.60 18572.0 26.65
7 Location_A II TR3 7160.50 42890.0 23.90
8 Location_A II TR4_5 3415.25 19067.0 29.00
9 Location_A III Control 7438.70 45417.0 20.80
10 Location_A III TR1_2 4808.60 24091.0 27.75
11 Location_A III TR3 6284.10 23713.0 23.90
12 Location_A III TR4_5 4275.55 23082.0 27.45
13 Location_A IV Control 5001.30 20770.0 20.80
14 Location_A IV TR1_2 4800.75 20763.5 28.95
15 Location_A IV TR3 7445.10 34173.0 20.90
16 Location_A IV TR4_5 5192.05 26227.0 27.70I want to summarize for each unique ID. So, I’ll add numbers from 1 up to the end.
dataA= dataA %>%
mutate(No=1:nrow(dataA))
dataA
Location Crop Plot Block Treatment Treatment_modified Total_DW Grain_yield AGW GN No
1 Location_A Sorghum 15 I Control Control 852.3 4303.6 28.1 31627 1
2 Location_A Sorghum 17 II Control Control 1295.7 6058.4 26.3 40157 2
3 Location_A Sorghum 19 III Control Control 1488.4 7438.7 20.8 45417 3
4 Location_A Sorghum 22 IV Control Control 925.6 5001.3 20.8 20770 4
5 Location_A Sorghum 1 I TR1 TR1_2 807.4 3285.2 30.7 14488 5
6 Location_A Sorghum 4 II TR1 TR1_2 907.4 3896.1 28.5 20323 6
7 Location_A Sorghum 9 III TR1 TR1_2 778.6 3207.3 28.8 15636 7
8 Location_A Sorghum 10 IV TR1 TR1_2 895.2 4418.2 28.6 20807 8
9 Location_A Sorghum 1 I TR2 TR1_2 748.9 3130.9 49.1 11556 9
10 Location_A Sorghum 4 II TR2 TR1_2 1032.4 4809.1 24.8 16821 10
11 Location_A Sorghum 9 III TR2 TR1_2 1388.2 6409.9 26.7 32546 11
12 Location_A Sorghum 10 IV TR2 TR1_2 1215.1 5183.3 29.3 20720 12
13 Location_A Sorghum 1 I TR3 TR3 668.7 3008.1 25.5 10390 13
14 Location_A Sorghum 4 II TR3 TR3 1564.1 7160.5 23.9 42890 14
15 Location_A Sorghum 9 III TR3 TR3 1283.4 6284.1 23.9 23713 15
16 Location_A Sorghum 10 IV TR3 TR3 1612.0 7445.1 20.9 34173 16
17 Location_A Sorghum 1 I TR4 TR4_5 1010.5 4340.3 29.2 23492 17
18 Location_A Sorghum 4 II TR4 TR4_5 757.4 2865.3 29.2 17564 18
19 Location_A Sorghum 9 III TR4 TR4_5 1087.1 5106.0 25.4 30184 19
20 Location_A Sorghum 10 IV TR4 TR4_5 1167.7 5383.6 27.7 23543 20
21 Location_A Sorghum 1 I TR5 TR4_5 783.3 2765.0 26.5 22890 21
22 Location_A Sorghum 4 II TR5 TR4_5 1002.5 3965.2 28.8 20570 22
23 Location_A Sorghum 9 III TR5 TR4_5 733.4 3445.1 29.5 15980 23
24 Location_A Sorghum 10 IV TR5 TR4_5 1067.9 5000.5 27.7 28911 24and I’ll summarize data again.
summary=data.frame(dataA %>%
group_by(No, Location,Block, Treatment_modified) %>%
dplyr::summarize(across(c(Grain_yield,GN,AGW),
.fns= list(Mean=~mean(., na.rm=TRUE)))))
summary
No Location Block Treatment_modified Grain_yield_Mean GN_Mean AGW_Mean
1 1 Location_A I Control 4303.6 31627 28.1
2 2 Location_A II Control 6058.4 40157 26.3
3 3 Location_A III Control 7438.7 45417 20.8
4 4 Location_A IV Control 5001.3 20770 20.8
5 5 Location_A I TR1_2 3285.2 14488 30.7
6 6 Location_A II TR1_2 3896.1 20323 28.5
7 7 Location_A III TR1_2 3207.3 15636 28.8
8 8 Location_A IV TR1_2 4418.2 20807 28.6
9 9 Location_A I TR1_2 3130.9 11556 49.1
10 10 Location_A II TR1_2 4809.1 16821 24.8
11 11 Location_A III TR1_2 6409.9 32546 26.7
12 12 Location_A IV TR1_2 5183.3 20720 29.3
13 13 Location_A I TR3 3008.1 10390 25.5
14 14 Location_A II TR3 7160.5 42890 23.9
15 15 Location_A III TR3 6284.1 23713 23.9
16 16 Location_A IV TR3 7445.1 34173 20.9
17 17 Location_A I TR4_5 4340.3 23492 29.2
18 18 Location_A II TR4_5 2865.3 17564 29.2
19 19 Location_A III TR4_5 5106.0 30184 25.4
20 20 Location_A IV TR4_5 5383.6 23543 27.7
21 21 Location_A I TR4_5 2765.0 22890 26.5
22 22 Location_A II TR4_5 3965.2 20570 28.8
23 23 Location_A III TR4_5 3445.1 15980 29.5
24 24 Location_A IV TR4_5 5000.5 28911 27.7Then, I’ll download this data to my PC.
library(writexl)
write_xlsx (summary,"C:/Users/Desktop/sorghum.xlsx")1) to upload data to SAS
First, let’s upload the data to SAS. I’ll assign this data to the Test table that I created.
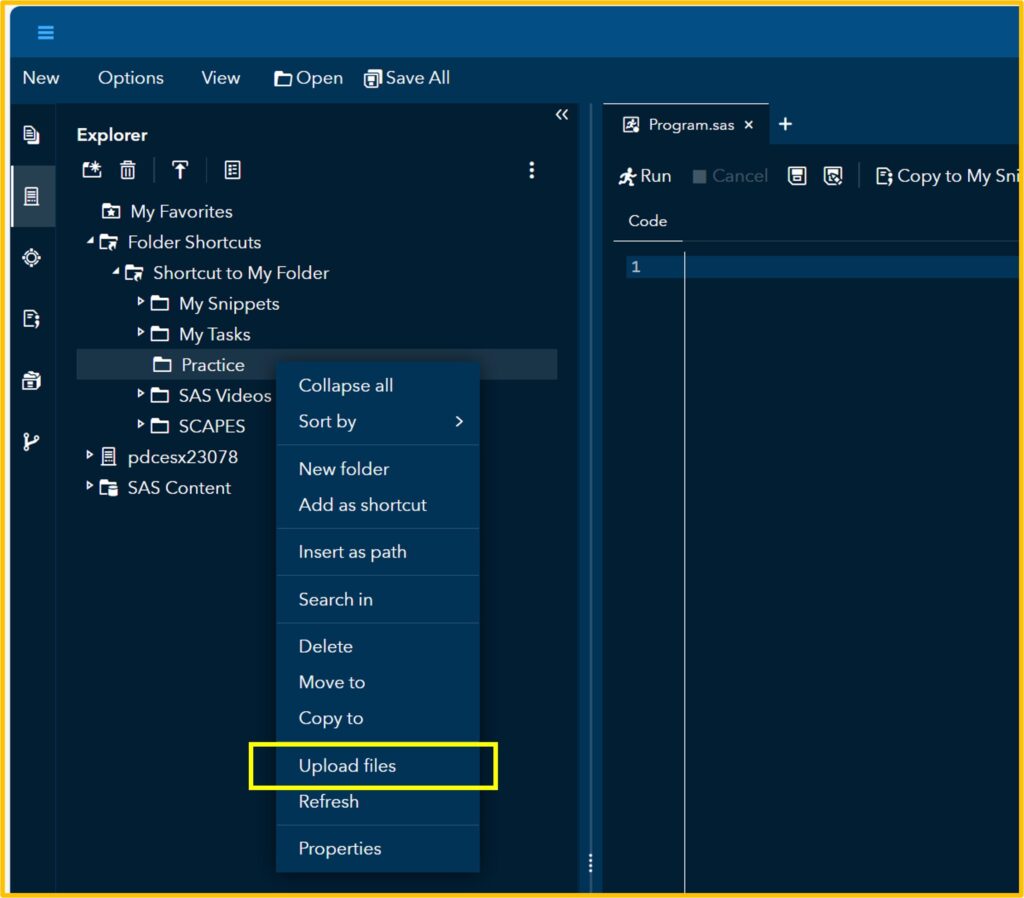
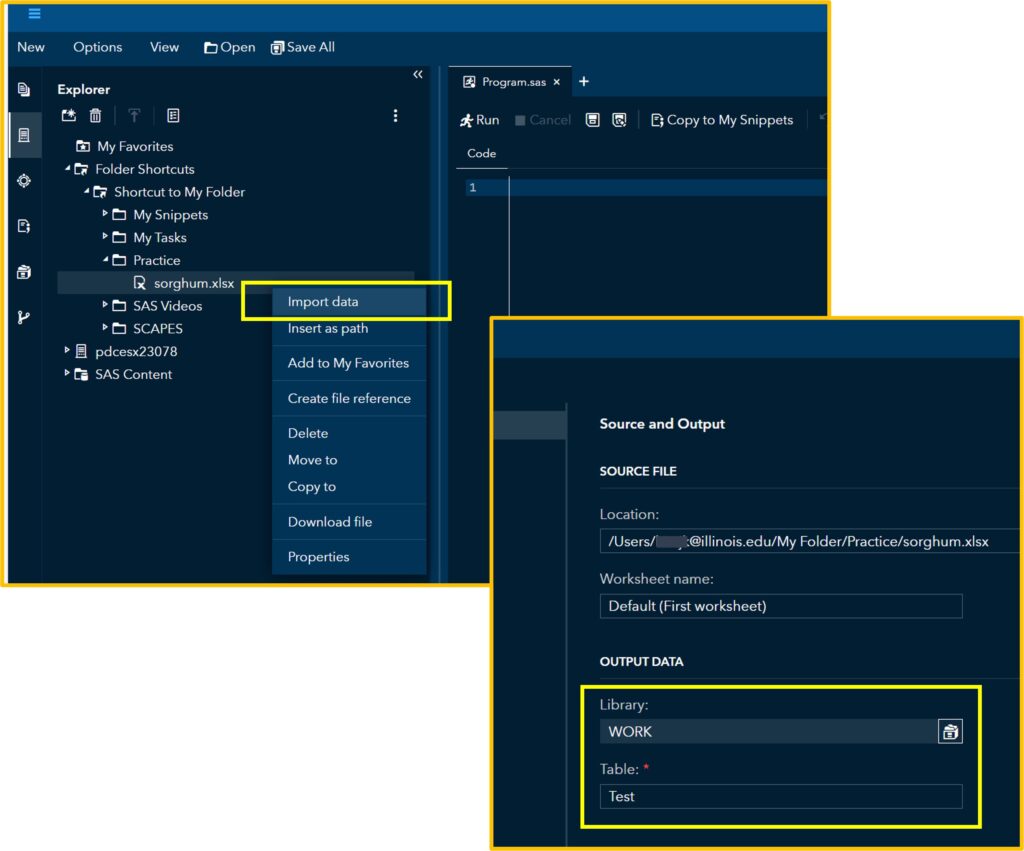
If you click Run, you can see the uploaded data in the Output Data. Then, click the Code tab.
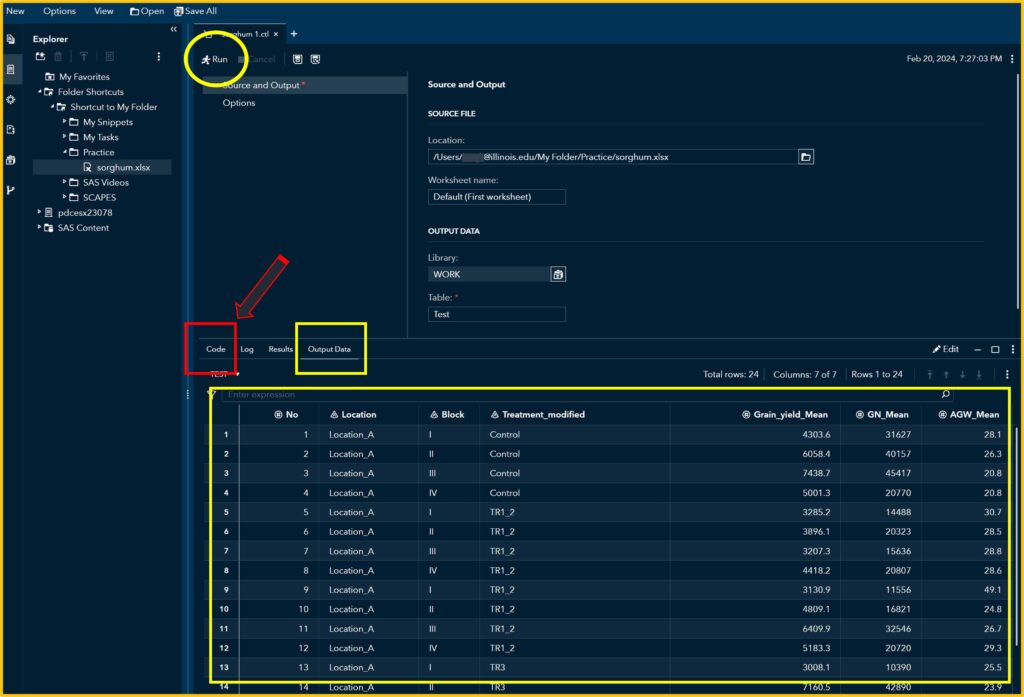
2) Coding
You can see the code that allows you to display the data. Click Edit.
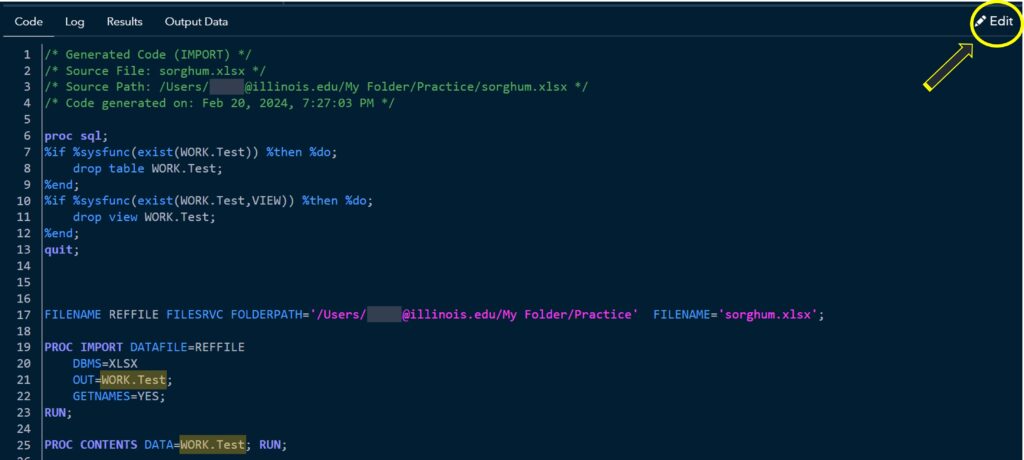
Then, you can add more codes. I’ll add the following codes:
proc sql;
create table Work.Control as
select *
from Work.Test
where Treatment_modified="Control";
quit;
PROC CONTENTS DATA=WORK.Control;
RUN;
proc sql;
create table Work.TR3 as
select *
from Work.Test
where Treatment_modified="TR3";
quit;
PROC CONTENTS DATA=WORK.TR3;
RUN;and run the code. You can find the newly created tables in the Libraries. In this code, I created two tables named Control and TR3, filtering specific variables.
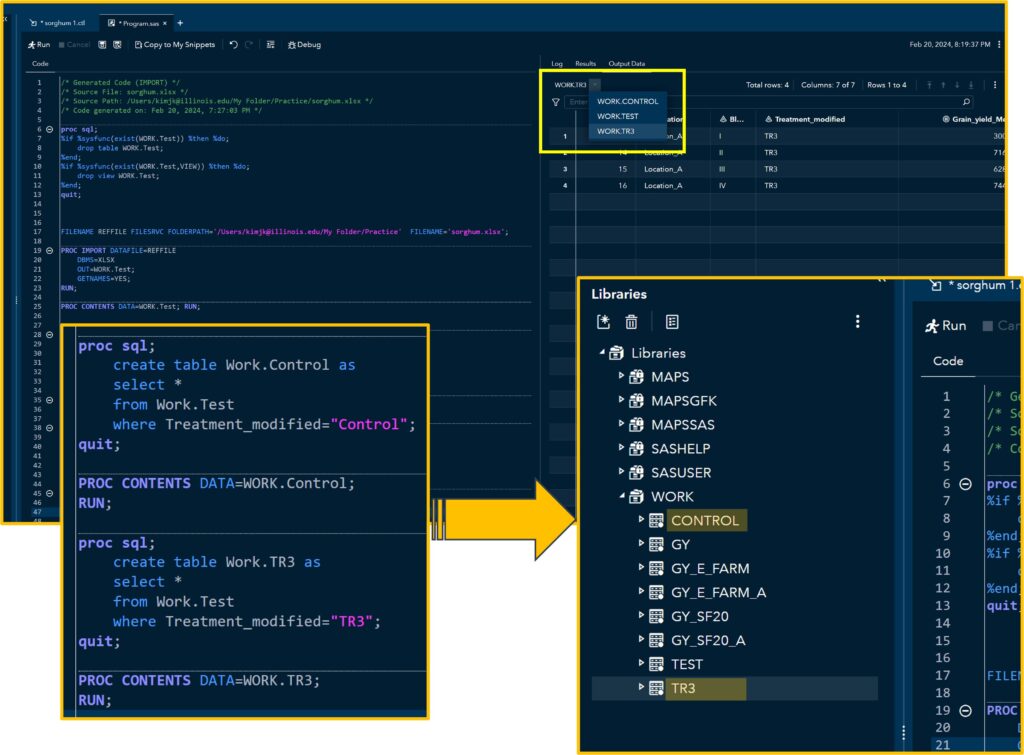
/* Generated Code (IMPORT) */
/* Source File: sorghum.xlsx */
/* Source Path: /Users/***@illinois.edu/My Folder/Practice/sorghum.xlsx */
/* Code generated on: Feb 20, 2024, 7:27:03 PM */
proc sql;
%if %sysfunc(exist(WORK.Test)) %then %do;
drop table WORK.Test;
%end;
%if %sysfunc(exist(WORK.Test,VIEW)) %then %do;
drop view WORK.Test;
%end;
quit;
FILENAME REFFILE FILESRVC FOLDERPATH='/Users/***@illinois.edu/My Folder/Practice' FILENAME='sorghum.xlsx';
PROC IMPORT DATAFILE=REFFILE
DBMS=XLSX
OUT=WORK.Test;
GETNAMES=YES;
RUN;
PROC CONTENTS DATA=WORK.Test; RUN;
proc sql;
create table Work.Control as
select *
from Work.Test
where Treatment_modified="Control";
quit;
PROC CONTENTS DATA=WORK.Control;
RUN;
proc sql;
create table Work.TR3 as
select *
from Work.Test
where Treatment_modified="TR3";
quit;
PROC CONTENTS DATA=WORK.TR3;
RUN;