How to add separate text to panels divided by facet_wrap() in R?
□ Graph Partitioning Using facet_wrap() in R Studio
□ How to customize the title format in facet_wrap()?
In my previous posts, I introduced how to divide panels in one figure using facet_wrap(). Today, I’ll introduce how to add separate text to panels.
First, let’s make sure we have the required packages installed.
library(dplyr)
library(tidyr)
library(ggplot2)I’ll create a dataset as shown below:
Nitrogen= c(rep("N0", 5), rep("N1", 5))
Cultivar_1= c(50,49,48,47,46,60,62,63,64,62)
Cultivar_2= c(55,57,56,55,54,65,66,67,64,63)
Cultivar_3= c(60,62,63,65,59,60,59,57,56,58)
dataA= data.frame(Nitrogen, Cultivar_1, Cultivar_2, Cultivar_3)
dataA
Nitrogen Cultivar_1 Cultivar_2 Cultivar_3
1 N0 50 55 60
2 N0 49 57 62
3 N0 48 56 63
4 N0 47 55 65
5 N0 46 54 59
6 N1 60 65 60
7 N1 62 66 59
8 N1 63 67 57
9 N1 64 64 56
10 N1 62 63 58Next, I’ll reshape the dataset into columns to facilitate data analysis.
df= data.frame(
dataA %>%
pivot_longer(
cols=c(Cultivar_1, Cultivar_2, Cultivar_3),
names_to="Genotype", values_to="Yield")
)
df
Nitrogen Genotype Yield
N0 Cultivar_1 50
N0 Cultivar_2 55
N0 Cultivar_3 60
N0 Cultivar_1 49
N0 Cultivar_2 57
N0 Cultivar_3 62
.
.
.And then, I’ll summarize this data using descriptive statistics.
dataB= data.frame(df %>%
group_by(Genotype, Nitrogen) %>%
dplyr::summarize(across(c(Yield),
.fns = list(Mean = mean,
SD = sd,
n = length,
se = ~ sd(.)/sqrt(length(.))))))
dataB
Genotype Nitrogen Yield_Mean Yield_SD Yield_n Yield_se
Cultivar_1 N0 48.0 1.581139 5 0.7071068
Cultivar_1 N1 62.2 1.483240 5 0.6633250
Cultivar_2 N0 55.4 1.140175 5 0.5099020
Cultivar_2 N1 65.0 1.581139 5 0.7071068
Cultivar_3 N0 61.8 2.387467 5 1.0677078
Cultivar_3 N1 58.0 1.581139 5 0.7071068
Finally, I’ll create a graph to visualize the summarized data.
ggplot (data=dataB, aes(x=Genotype, y=Yield_Mean, fill=Nitrogen)) +
geom_bar(stat="identity",position="dodge") +
geom_errorbar(aes(ymin= Yield_Mean-Yield_se, ymax= Yield_Mean+Yield_se),
position=position_dodge(0.9), width=0.2) +
scale_fill_manual(values= c("Gray","Dark green")) +
scale_y_continuous(breaks= seq(0, 100, 20), limits= c(0, 100))+
facet_wrap(~Nitrogen) +
labs(x="Cultivar", y="Yield") +
theme_classic(base_size=18, base_family="serif")+
theme(legend.position='none',
legend.title=element_blank(),
legend.key=element_rect(color="white", fill="white"),
legend.text=element_text(family="serif", face="plain",
size=15, color= "Black"),
legend.background=element_rect(fill="white"),
axis.line=element_line(linewidth=0.5, colour="black"),
strip.background=element_rect(color="white",
linewidth=0.5,linetype="solid"))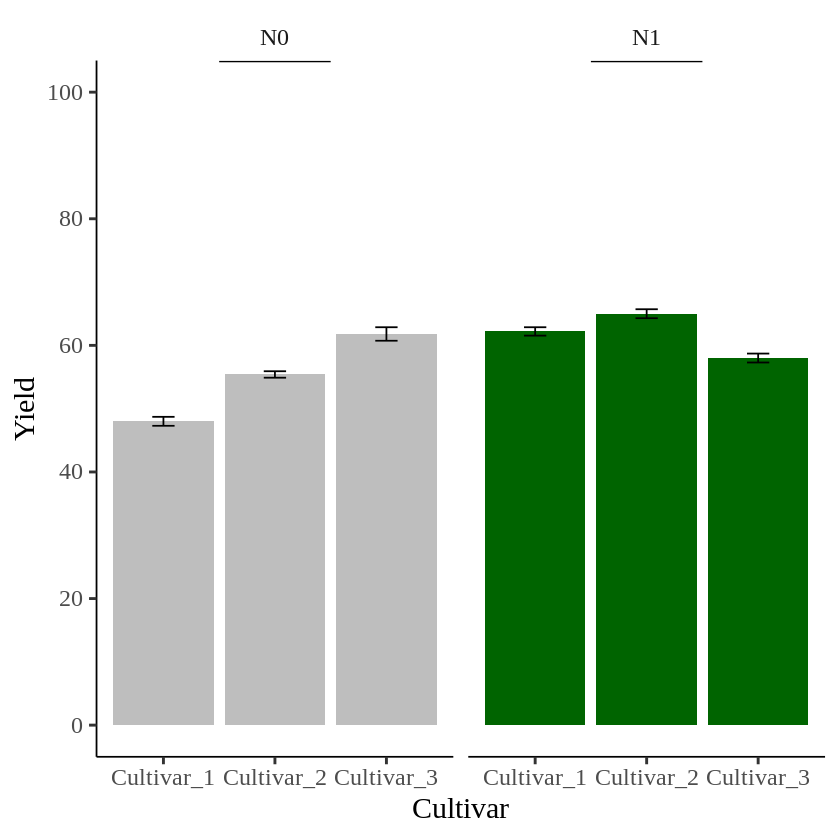
In this graph, I want to indicate statistical significance with asterisks (***) in each panel. First, I’ll add asterisks in the N0 panel.
ggplot (data=dataB, aes(x=Genotype, y=Yield_Mean, fill=Nitrogen)) +
geom_bar(stat="identity",position="dodge") +
geom_errorbar(aes(ymin= Yield_Mean-Yield_se, ymax= Yield_Mean+Yield_se),
position=position_dodge(0.9), width=0.2) +
scale_fill_manual(values= c("Gray","Dark green")) +
###
geom_text(aes(family="serif",fontface=6), x=1.5, y=80, label="***", size=7, col="red") +
###
scale_y_continuous(breaks= seq(0, 100, 20), limits= c(0, 100))+
facet_wrap(~Nitrogen) +
annotate("segment", x=1.5, xend=2.5, y=Inf,yend=Inf, color="black", lwd=1)+
labs(x="Cultivar", y="Yield") +
theme_classic(base_size=18, base_family="serif")+
theme(legend.position='none',
legend.title=element_blank(),
legend.key=element_rect(color="white", fill="white"),
legend.text=element_text(family="serif", face="plain",
size=15, color= "Black"),
legend.background=element_rect(fill="white"),
axis.line=element_line(linewidth=0.5, colour="black"),
strip.background=element_rect(color="white",
linewidth=0.5,linetype="solid"))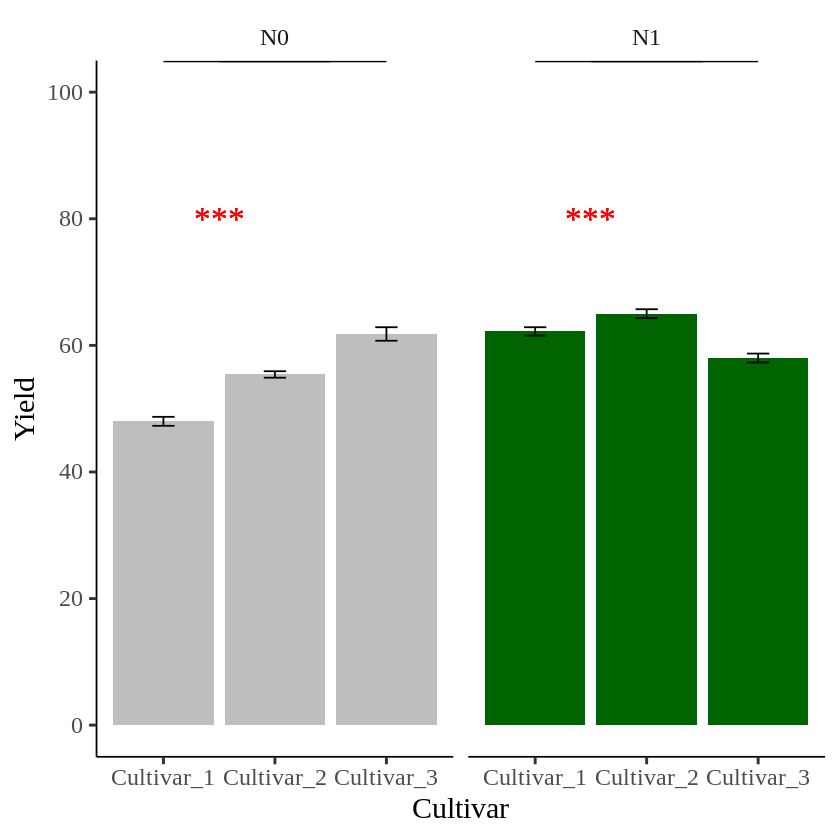
The problem is that *** is added to both panels. I want a different symbol for each panel.
to create data table to insert text to specific panel
I’ll create a data table.
text1= data.frame(Nitrogen="N0", STAT= "***", Genotype="Cultivar_2", Yield=70)
text1
Nitrogen STAT Genotype Yield
N0 *** Cultivar_2 70
I’ll add this data table to the geom_text().
geom_text(data=text1, aes(family="serif", x=Genotype, y=Yield, label=STAT), size=5, color="red")Let’s create a graph again, this time to visualize the data.
ggplot (data=dataB, aes(x=Genotype, y=Yield_Mean, fill=Nitrogen)) +
geom_bar(stat="identity",position="dodge") +
geom_errorbar(aes(ymin= Yield_Mean-Yield_se, ymax= Yield_Mean+Yield_se),
position=position_dodge(0.9), width=0.2) +
scale_fill_manual(values= c("Gray","Dark green")) +
###
geom_text(data=text1, aes(family="serif", x=Genotype, y=Yield, label=STAT), size=5, color="red") +
###
scale_y_continuous(breaks= seq(0, 100, 20), limits= c(0, 100))+
facet_wrap(~Nitrogen) +
annotate("segment", x=1.5, xend=2.5, y=Inf,yend=Inf, color="black", lwd=1)+
labs(x="Cultivar", y="Yield") +
theme_classic(base_size=18, base_family="serif")+
theme(legend.position='none',
legend.title=element_blank(),
legend.key=element_rect(color="white", fill="white"),
legend.text=element_text(family="serif", face="plain",
size=15, color= "Black"),
legend.background=element_rect(fill="white"),
axis.line=element_line(linewidth=0.5, colour="black"),
strip.background=element_rect(color="white",
linewidth=0.5,linetype="solid"))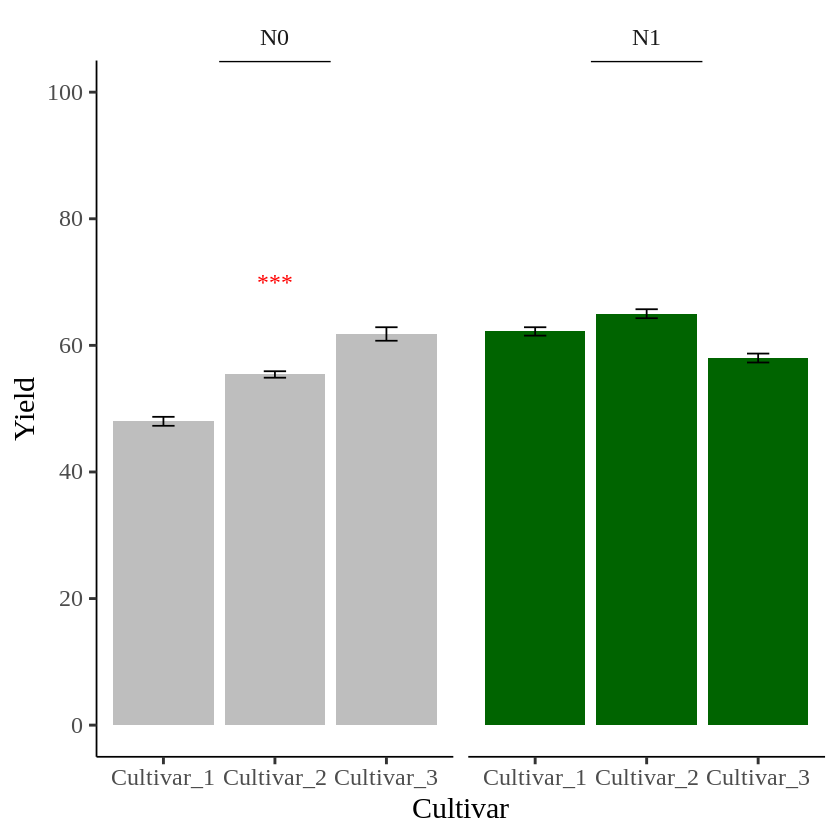
I’ll create another data table to add text in the second panel.
text2= data.frame(Nitrogen="N1", STAT= "**", Genotype="Cultivar_1", Yield=80)
geom_text(data=text2, aes(family="serif", x=Genotype, y=Yield, label=STAT), size=5, color="red")
##
ggplot (data=dataB, aes(x=Genotype, y=Yield_Mean, fill=Nitrogen)) +
geom_bar(stat="identity",position="dodge") +
geom_errorbar(aes(ymin= Yield_Mean-Yield_se, ymax= Yield_Mean+Yield_se),
position=position_dodge(0.9), width=0.2) +
scale_fill_manual(values= c("Gray","Dark green")) +
###
geom_text(data=text1, aes(family="serif", x=Genotype, y=Yield, label=STAT), size=5, color="red") +
geom_text(data=text2, aes(family="serif", x=Genotype, y=Yield, label=STAT), size=5, color="red") +
###
scale_y_continuous(breaks= seq(0, 100, 20), limits= c(0, 100))+
facet_wrap(~Nitrogen) +
annotate("segment", x=1.5, xend=2.5, y=Inf,yend=Inf, color="black", lwd=1)+
labs(x="Cultivar", y="Yield") +
theme_classic(base_size=18, base_family="serif")+
theme(legend.position='none',
legend.title=element_blank(),
legend.key=element_rect(color="white", fill="white"),
legend.text=element_text(family="serif", face="plain",
size=15, color= "Black"),
legend.background=element_rect(fill="white"),
axis.line=element_line(linewidth=0.5, colour="black"),
strip.background=element_rect(color="white",
linewidth=0.5,linetype="solid"))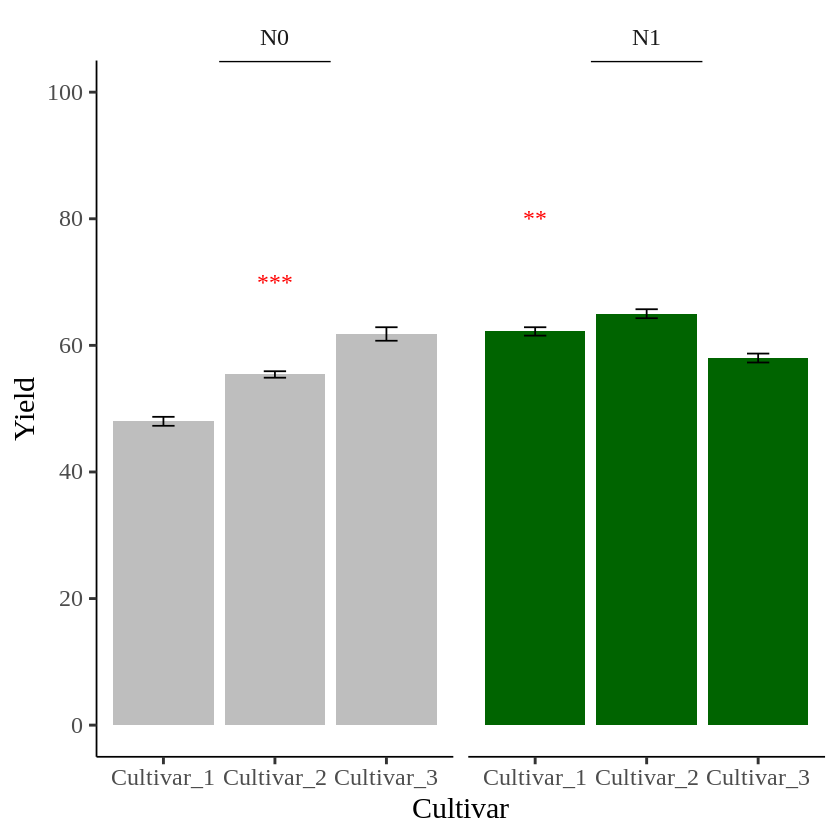
Now, different asterisks (***) are indicated in each panel.
full code: https://github.com/agronomy4future/r_code/blob/main/How_to_add_separate_text_to_panels_divided_by_facet_wrap()_in_R.ipynb
