Utilizing R Studio for Data Grouping and Mean/Standard Error Calculation (feat ddply)
The function I will introduce today is ddply(). This function is convenient for summarizing large amounts of data and can also calculate standard errors, making it easy to create bar graphs. First, install the package.
install.packages ("plyr")
library(plyr)Once the installation is complete, let’s upload some data.
#install.packages ("readr")
library(readr)
github= "https://raw.githubusercontent.com/agronomy4future/raw_data_practice/main/fertilizer_treatment.csv"
dataA= data.frame(read_csv(url(github), show_col_types = FALSE))
print(dataA)
Genotype Block variable value
1 Genotype_A I Control 42.9
2 Genotype_A II Control 41.6
3 Genotype_A III Control 28.9
4 Genotype_A IV Control 30.8
5 Genotype_B I Control 53.3
6 Genotype_B II Control 69.6
7 Genotype_B III Control 45.4
8 Genotype_B IV Control 35.1
9 Genotype_C I Control 62.3
10 Genotype_C II Control 58.5
11 Genotype_C III Control 44.6
12 Genotype_C IV Control 50.3
13 Genotype_D I Control 75.4
14 Genotype_D II Control 65.6
15 Genotype_D III Control 54.0
16 Genotype_D IV Control 52.7
17 Genotype_A I Fertilizer1 53.8
18 Genotype_A II Fertilizer1 58.5
19 Genotype_A III Fertilizer1 43.9
20 Genotype_A IV Fertilizer1 46.3
21 Genotype_B I Fertilizer1 57.6
22 Genotype_B II Fertilizer1 69.6
23 Genotype_B III Fertilizer1 42.4
24 Genotype_B IV Fertilizer1 51.9
25 Genotype_C I Fertilizer1 63.4
26 Genotype_C II Fertilizer1 50.4
27 Genotype_C III Fertilizer1 45.0
28 Genotype_C IV Fertilizer1 46.7
29 Genotype_D I Fertilizer1 70.3
30 Genotype_D II Fertilizer1 67.3
31 Genotype_D III Fertilizer1 57.6
32 Genotype_D IV Fertilizer1 58.5
33 Genotype_A I Fertilizer2 49.5
34 Genotype_A II Fertilizer2 53.8
35 Genotype_A III Fertilizer2 40.7
36 Genotype_A IV Fertilizer2 39.4
37 Genotype_B I Fertilizer2 59.8
38 Genotype_B II Fertilizer2 65.8
39 Genotype_B III Fertilizer2 41.4
40 Genotype_B IV Fertilizer2 45.4
41 Genotype_C I Fertilizer2 64.5
42 Genotype_C II Fertilizer2 46.1
43 Genotype_C III Fertilizer2 62.6
44 Genotype_C IV Fertilizer2 50.3
45 Genotype_D I Fertilizer2 68.8
46 Genotype_D II Fertilizer2 65.3
47 Genotype_D III Fertilizer2 45.6
48 Genotype_D IV Fertilizer2 51.0
49 Genotype_A I Fertilizer3 44.4
50 Genotype_A II Fertilizer3 41.8
51 Genotype_A III Fertilizer3 28.3
52 Genotype_A IV Fertilizer3 34.7
53 Genotype_B I Fertilizer3 64.1
54 Genotype_B II Fertilizer3 57.4
55 Genotype_B III Fertilizer3 44.1
56 Genotype_B IV Fertilizer3 51.6
57 Genotype_C I Fertilizer3 63.6
58 Genotype_C II Fertilizer3 56.1
59 Genotype_C III Fertilizer3 52.7
60 Genotype_C IV Fertilizer3 51.8
61 Genotype_D I Fertilizer3 71.6
62 Genotype_D II Fertilizer3 69.4
63 Genotype_D III Fertilizer3 56.6
64 Genotype_D IV Fertilizer3 47.4This dataset consists of results from cultivating 4 genotypes under 4 different nitrogen treatment conditions with 4 replicates each. In other words, it comprises a total of 64 data points (4 x 4 x 4). When summarizing this data, you can utilize the ddply() function. Now, I’d like to summarize this data by calculating the mean, standard deviation, and standard error. Please write the code in the following format:
dataB= ddply (dataA,c("Genotype","variable"), summarise, mean=mean(value),
sd=sd(value),n=length(value), se=sd/sqrt(n))
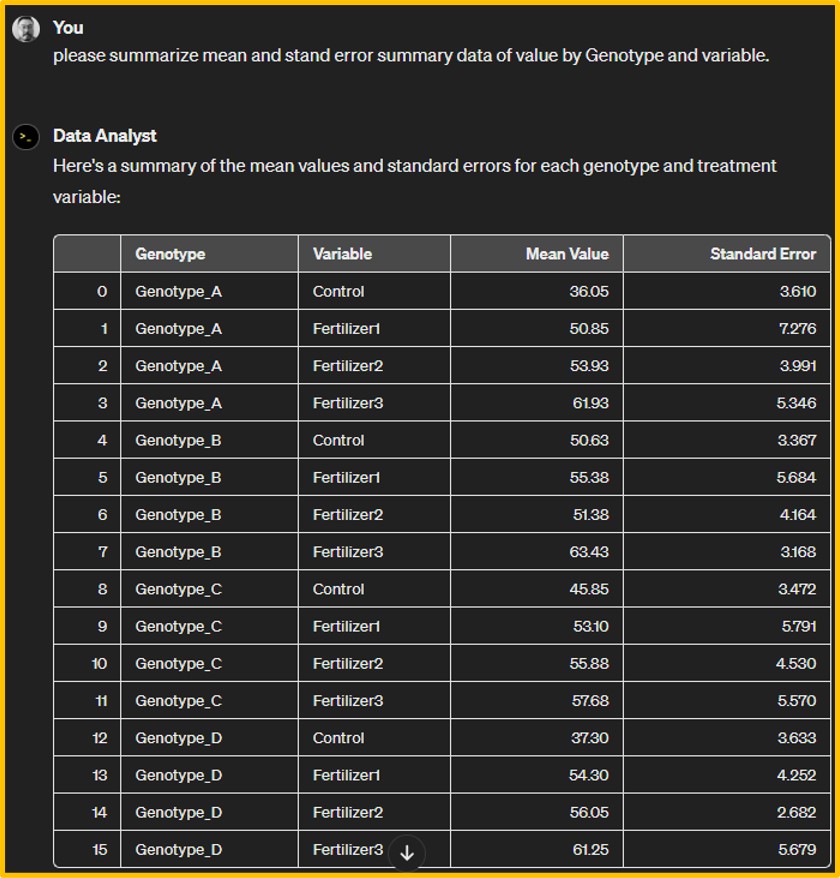
Genotype variable mean sd n se
1 Genotype_A Control 36.050 7.220572 4 3.610286
2 Genotype_A Fertilizer1 50.625 6.733684 4 3.366842
3 Genotype_A Fertilizer2 45.850 6.943822 4 3.471911
4 Genotype_A Fertilizer3 37.300 7.266820 4 3.633410
5 Genotype_B Control 50.850 14.552548 4 7.276274
6 Genotype_B Fertilizer1 55.375 11.368487 4 5.684244
7 Genotype_B Fertilizer2 53.100 11.581019 4 5.790509
8 Genotype_B Fertilizer3 54.300 8.504509 4 4.252254
9 Genotype_C Control 53.925 7.982637 4 3.991319
10 Genotype_C Fertilizer1 51.375 8.327615 4 4.163807
11 Genotype_C Fertilizer2 55.875 9.059939 4 4.529970
12 Genotype_C Fertilizer3 56.050 5.363146 4 2.681573
13 Genotype_D Control 61.925 10.692482 4 5.346241
14 Genotype_D Fertilizer1 63.425 6.336863 4 3.168432
15 Genotype_D Fertilizer2 57.675 11.139532 4 5.569766
16 Genotype_D Fertilizer3 61.250 11.357670 4 5.678835The data was organized into means, standard deviations, and standard errors based on 4 replicates for both the variety and treatment. If the fertilizer treatment does not hold significant meaning, we could also organize the data by grouping only by genotype.
dataC= ddply (dataA,c("Genotype"), summarise, mean=mean(value),
sd=sd(value),n=length(value), se=sd/sqrt(n))
Genotype mean sd n se
1 Genotype_A 42.45625 8.865662 16 2.216415
2 Genotype_B 53.40625 10.606945 16 2.651736
3 Genotype_C 54.30625 7.251250 16 1.812812
4 Genotype_D 61.06875 9.288180 16 2.322045However, ddply() is considered outdated code!!
Nowadays, no one uses ddply() because there are more convenient methods available. I will introduce the simplest method to summarize data using dplyr().
library(dplyr)
dataB= data.frame(dataA %>%
group_by(Genotype, variable) %>%
dplyr::summarize(across(c(value),
.fns = list(Mean = mean,
SD = sd,
n = length,
se = ~ sd(.)/sqrt(length(.))))))
dataB
Genotype variable value_Mean value_SD value_n value_se
1 Genotype_A Control 36.050 7.220572 4 3.610286
2 Genotype_A Fertilizer1 50.625 6.733684 4 3.366842
3 Genotype_A Fertilizer2 45.850 6.943822 4 3.471911
4 Genotype_A Fertilizer3 37.300 7.266820 4 3.633410
5 Genotype_B Control 50.850 14.552548 4 7.276274
6 Genotype_B Fertilizer1 55.375 11.368487 4 5.684244
7 Genotype_B Fertilizer2 53.100 11.581019 4 5.790509
8 Genotype_B Fertilizer3 54.300 8.504509 4 4.252254
9 Genotype_C Control 53.925 7.982637 4 3.991319
10 Genotype_C Fertilizer1 51.375 8.327615 4 4.163807
11 Genotype_C Fertilizer2 55.875 9.059939 4 4.529970
12 Genotype_C Fertilizer3 56.050 5.363146 4 2.681573
13 Genotype_D Control 61.925 10.692482 4 5.346241
14 Genotype_D Fertilizer1 63.425 6.336863 4 3.168432
15 Genotype_D Fertilizer2 57.675 11.139532 4 5.569766
16 Genotype_D Fertilizer3 61.250 11.357670 4 5.678835Tip) AI can easily summarizes data!!
These days we do not have to learn programming code because AI simply summarize the data if we just ask.
ChatGPT - Data Analyst
You can upload the data file to Data Analyst, but I’ll upload code to generate data table. If you run the following code, you can obtain the data frame code to generate data table.
library(readr)
github= "https://raw.githubusercontent.com/agronomy4future/raw_data_practice/main/fertilizer_treatment.csv"
dataA= data.frame(read_csv(url(github), show_col_types = FALSE))
print(dataA)
#remotes::install_github("cynkra/constructive")
library(remote)
constructive::construct(dataA)
data.frame(
Genotype= rep(rep(c("Genotype_A", "Genotype_B", "Genotype_C", "Genotype_D"), 4), each = 4L),
Block= rep(c("I", "II", "III", "IV"), 16),
variable= rep(c("Control", "Fertilizer1", "Fertilizer2", "Fertilizer3"), each= 16L),
value= c(42.9, 41.6, 28.9, 30.8, 53.3, 69.6, 45.4, 35.1, 62.3, 58.5, 44.6, 50.3, 75.4, 65.6, 54, 52.7, 53.8, 58.5, 43.9, 46.3, 57.6, 69.6, 42.4, 51.9, 63.4, 50.4, 45, 46.7, 70.3, 67.3, 57.6, 58.5, 49.5, 53.8, 40.7, 39.4, 59.8, 65.8, 41.4, 45.4, 64.5, 46.1, 62.6, 50.3, 68.8, 65.3, 45.6, 51, 44.4, 41.8, 28.3, 34.7, 64.1, 57.4, 44.1, 51.6, 63.6, 56.1, 52.7, 51.8, 71.6, 69.4, 56.6, 47.4)
)I typed the code (blue) to create data, and asked ChatGPT to summarize mean summary data with standard error.