Creating Stacked Bar Graphs in R Studio: A Step-by-Step Guide
Today, I’ll be introducing how to create stacked bar graphs using R Studio. To start, I will generate a data table as shown below.
Genotype= c(rep(c("CV1","CV2", "CV3"), each=9))
Treatment= c(rep(c("TR1", "TR2", "TR3"), each=3),
rep(c("TR1", "TR2", "TR3"), each=3),
rep(c("TR1", "TR2", "TR3"), each=3))
Block= c(rep(c("B1","B2","B3"), times=9))
yield= c(rep("15",5), rep("18",5), rep("20",8), rep("14",7), rep ("21",2))
dataA= data.frame (Genotype, Treatment, Block, yield)
Genotype Treatment Block yield
1 CV1 TR1 B1 15
2 CV1 TR1 B2 15
3 CV1 TR1 B3 15
4 CV1 TR2 B1 15
5 CV1 TR2 B2 15
6 CV1 TR2 B3 18
7 CV1 TR3 B1 18
8 CV1 TR3 B2 18
9 CV1 TR3 B3 18
10 CV2 TR1 B1 18
11 CV2 TR1 B2 20
12 CV2 TR1 B3 20
13 CV2 TR2 B1 20
14 CV2 TR2 B2 20
15 CV2 TR2 B3 20
16 CV2 TR3 B1 20
17 CV2 TR3 B2 20
18 CV2 TR3 B3 20
19 CV3 TR1 B1 14
20 CV3 TR1 B2 14
21 CV3 TR1 B3 14
22 CV3 TR2 B1 14
23 CV3 TR2 B2 14
24 CV3 TR2 B3 14
25 CV3 TR3 B1 14
26 CV3 TR3 B2 21
27 CV3 TR3 B3 21I’ll make stacked bar graphs using this data table. First of all, it’s necessary to summarize the data. I’ll use ddply() function.
install.packages("plyr")
library (plyr)
dataA_summary= ddply (dataA, c('Genotype','Treatment'), summarise, mean=mean(yield), sd=sd(yield), n=length(yield), se=sd/sqrt(n))If I use this code, the error message pops up
In mean.default(yield) :
argument is not numeric or logical: returning NAThis is because when generating data, I used double quotation marks such as yield = c(rep("15", 5), rep("18", 5), rep("20", 8), rep("14", 7), rep("21", 2)), causing R to interpret this data as text. Therefore, it’s necessary to explicitly convert these values to numbers using as.numeric(yield). If you wish to avoid this step, refrain from using double quotation marks when generating the yield variable.
dataA_summary= ddply (dataA, c('Genotype','Treatment'), summarise, mean=mean(as.numeric(yield)),sd=sd(as.numeric(yield)), n=length(as.numeric(yield)), se=sd/sqrt(n))
Genotype Treatment mean sd n se
1 CV1 TR1 15.00000 0.000000 3 0.0000000
2 CV1 TR2 16.00000 1.732051 3 1.0000000
3 CV1 TR3 18.00000 0.000000 3 0.0000000
4 CV2 TR1 19.33333 1.154701 3 0.6666667
5 CV2 TR2 20.00000 0.000000 3 0.0000000
6 CV2 TR3 20.00000 0.000000 3 0.0000000
7 CV3 TR1 14.00000 0.000000 3 0.0000000
8 CV3 TR2 14.00000 0.000000 3 0.0000000
9 CV3 TR3 18.66667 4.041452 3 2.3333333If you run the code, you will obtain this summarized data. This data includes the mean, standard deviation, and standard error. Now, let’s proceed to create a bar graph.
library (ggplot2)
ggplot(data=dataA_summary, aes(x=Treatment, y=mean, fill=Genotype))+
geom_bar(stat="identity",position="dodge", width= 0.7, size=1) +
geom_errorbar(aes(ymin= mean-se, ymax=mean + se), position=position_dodge(0.7),
width=0.2, color='Black') +
scale_fill_manual(values= c ("Cadetblue", "Dark gray","Dark orange")) +
scale_y_continuous(breaks = seq(0,25,5), limits= c(0,25)) +
labs(fill = "Genotype", x="Treatment", y="Yield") +
theme(axis.title= element_text (face= "plain", size= 15, color= "black"),
axis.text.x= element_text(size= 12),
axis.text.y= element_text(size= 15),
axis.line= element_line(size = 0.5, colour= "black"),
legend.position= 'right',
legend.key= element_rect(color= "white", fill= "white"),
legend.key.size= unit(0.5,"cm"),
legend.title= element_text(face= "plain", size= 12, color= "Black"),
legend.text= element_text(face= "plain", size= 12, color= "Black"),
strip.text.x= element_text(size= 15)) +
windows(width=5.5, height=5)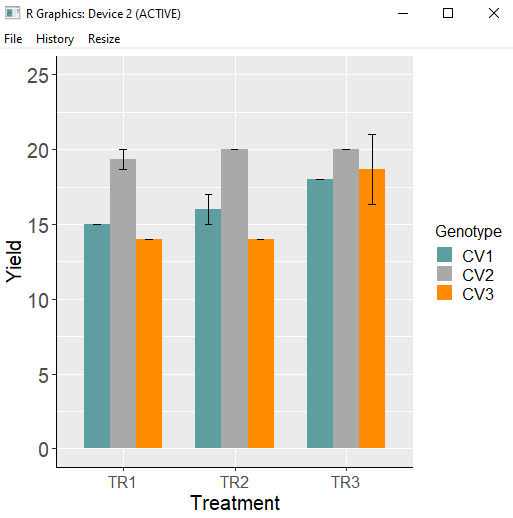
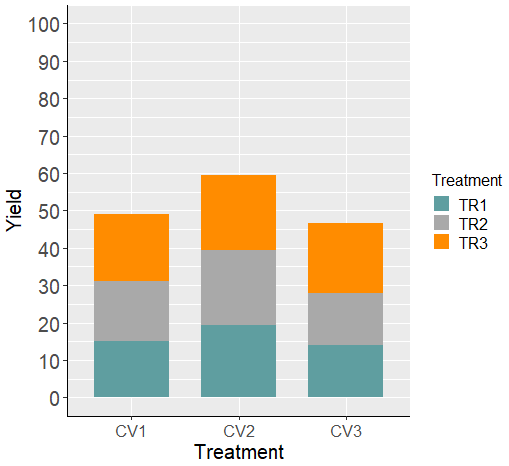
We have created a bar graph. However, now I want to create a stacked bar graph by stacking TR1 to TR3. In this case, we can achieve this by simply modifying the code: change position="dodge" to position="stack".
Furthermore, the x-axis should be labeled as “Genotype” because I aim to create a stacked bar graph based on TR per Genotype. Moreover, I’ve updated the legend’s name to “Treatment.”
ggplot(data=dataA_summary, aes(x=Genotype, y=mean, fill=Treatment))+
geom_bar(stat="identity", position = 'stack', width= 0.7, size=1) +
scale_fill_manual(values= c ("Cadetblue", "Dark gray","Dark orange")) +
scale_y_continuous(breaks= seq(0,100,10), limits= c(0,100)) +
labs(fill= "Treatment", x="Treatment", y="Yield") +
theme(axis.title= element_text (face= "plain", size = 15, color = "black"),
axis.text.x= element_text(size= 12),
axis.text.y= element_text(size= 15),
axis.line= element_line(size= 0.5, colour= "black"),
legend.position= 'right',
legend.key= element_rect(color= "white", fill= "white"),
legend.key.size= unit(0.5,"cm"),
legend.title= element_text(face= "plain", size= 12, color= "Black"),
legend.text= element_text(face= "plain", size= 12, color= "Black"),
strip.text.x= element_text(size= 15)) +
windows(width=5.5, height=5)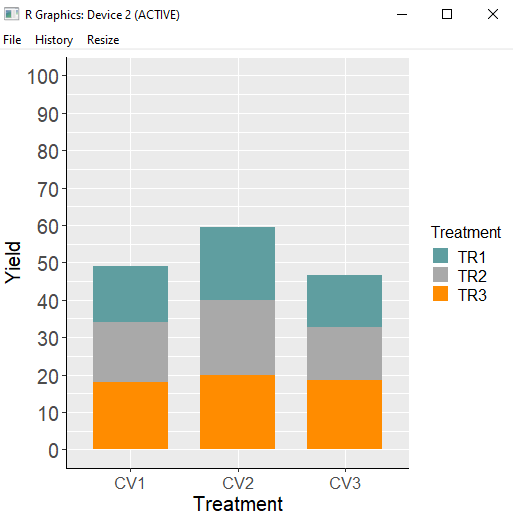
If you want to reverse the order of TR on the stacked bar graph, you can achieve that by adding the following code: position = position_stack(reverse = TRUE).
ggplot(data=dataA_summary, aes(x=Genotype, y=mean, fill=Treatment))+
geom_bar(stat="identity", position = position_stack(reverse=T), width = 0.7, size=1) +
scale_fill_manual(values= c ("Cadetblue", "Dark gray","Dark orange")) +
scale_y_continuous(breaks = seq(0,100,10), limits = c(0,100)) +
labs(fill = "Treatment", x="Treatment", y="Yield") +
theme(axis.title = element_text (face = "plain", size = 15, color = "black"),
axis.text.x = element_text(size= 12),
axis.text.y = element_text(size= 15),
axis.line = element_line(size = 0.5, colour = "black"),
legend.position = 'right',
legend.key = element_rect(color = "white", fill = "white"),
legend.key.size = unit(0.5,"cm"),
legend.title = element_text(face = "plain", size = 12, color = "Black"),
legend.text = element_text(face = "plain", size = 12, color = "Black"),
strip.text.x = element_text(size = 15)) +
windows(width=5.5, height=5)