Graph Partitioning Using facet_wrap() in R Studio
While creating graphs, you can certainly draw multiple graphs in a single panel. However, you can also use the facet_wrap() function to divide graphs based on specific variables.
First, let’s generate a dataset.
Nitrogen= c(rep("N0", 5), rep("N1", 5))
Cultivar_1= c(50,49,48,47,46,60,62,63,64,62)
Cultivar_2= c(55,57,56,55,54,65,66,67,64,63)
Cultivar_3= c(60,62,63,65,59,60,59,57,56,58)
dataA= data.frame(Nitrogen, Cultivar_1, Cultivar_2, Cultivar_3)
Nitrogen Cultivar_1 Cultivar_2 Cultivar_3
1 N0 50 55 60
2 N0 49 57 62
3 N0 48 56 63
4 N0 47 55 65
5 N0 46 54 59
6 N1 60 65 60
7 N1 62 66 59
8 N1 63 67 57
9 N1 64 64 56
10 N1 62 63 58I intend to create a bar graph using this data. Therefore, I need to summarize the data. To do this, I must reorganize the data from being divided into columns to being arranged in rows.
dataB= reshape2::melt(dataA[c("Nitrogen","Cultivar_1","Cultivar_2", "Cultivar_3")], id.vars=c("Nitrogen"))
Nitrogen variable value
1 N0 Cultivar_1 50
2 N0 Cultivar_1 49
3 N0 Cultivar_1 48
4 N0 Cultivar_1 47
5 N0 Cultivar_1 46
6 N1 Cultivar_1 60
7 N1 Cultivar_1 62
8 N1 Cultivar_1 63
9 N1 Cultivar_1 64
10 N1 Cultivar_1 62
11 N0 Cultivar_2 55
12 N0 Cultivar_2 57
13 N0 Cultivar_2 56
14 N0 Cultivar_2 55
15 N0 Cultivar_2 54
16 N1 Cultivar_2 65
17 N1 Cultivar_2 66
18 N1 Cultivar_2 67
19 N1 Cultivar_2 64
20 N1 Cultivar_2 63
21 N0 Cultivar_3 60
22 N0 Cultivar_3 62
23 N0 Cultivar_3 63
24 N0 Cultivar_3 65
25 N0 Cultivar_3 59
26 N1 Cultivar_3 60
27 N1 Cultivar_3 59
28 N1 Cultivar_3 57
29 N1 Cultivar_3 56
30 N1 Cultivar_3 58I have reorganized the data into rows using the reshape2::melt() function. Now, let’s proceed to summarize this data. I will be using the dplyr package for this purpose.
library (dplyr)
dataC= data.frame(dataB %>%
group_by(Nitrogen, variable) %>%
summarise(mean=mean(value), sd=sd(value), n=length(value), se=sd/sqrt(n)))
Nitrogen variable mean sd n se
1 N0 Cultivar_1 48.0 1.581139 5 0.7071068
2 N0 Cultivar_2 55.4 1.140175 5 0.5099020
3 N0 Cultivar_3 61.8 2.387467 5 1.0677078
4 N1 Cultivar_1 62.2 1.483240 5 0.6633250
5 N1 Cultivar_2 65.0 1.581139 5 0.7071068
6 N1 Cultivar_3 58.0 1.581139 5 0.7071068The data has been summarized as follows: The average and standard error of the yield for three varieties based on Nitrogen treatment (N0, N1) have been summarized. With this data, let’s proceed to create a bar graph.
library(ggplot2)
ggplot (data=dataC, aes(x=variable, y=mean, fill=Nitrogen)) +
geom_bar(stat="identity",position="dodge") +
geom_errorbar(aes(ymin= mean-se, ymax=mean+se), position=position_dodge(0.9),
width=0.2) +
scale_y_continuous(breaks= seq(0, 70, 10), limits= c(0, 70))+
scale_fill_manual(values= c("Gray","Dark green")) +
labs(fill="Nitrogen", x="Cultivar", y="Yield") +
theme(axis.title= element_text (face= "plain", size= 17, color= "black"),
axis.text.x= element_text(size= 15),
axis.text.y= element_text(size= 15),
axis.line= element_line(size= 0.5, colour= "black"),
legend.position= 'bottom',
legend.key= element_rect(color= "white", fill= "white"),
legend.key.size= unit(0.5,"cm"),
legend.title= element_text(face="plain", size= 14, color= "Black"),
legend.text= element_text(face="plain", size= 14, color= "Black"),
strip.text.x= element_text(size=17),
plot.margin=unit(c(0.5,0.5,0.5,0.5),"cm")) +
windows(width=5.5, height=5)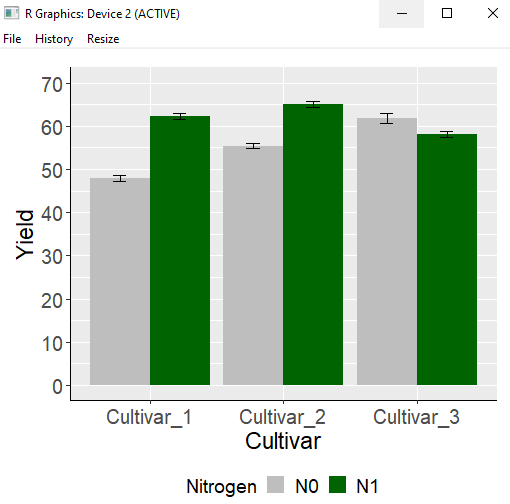
I have created the bar graph. I used aes(x=variable, y=mean, fill=Nitrogen) and assigned Nitrogen to the fill aesthetic. As a result, in a single graph panel, two bar graphs are displayed for each cultivar based on the Nitrogen factor. However, instead of this approach, I would like to create individual bar graphs for each Nitrogen level. Rather than plotting graphs in separate panels, I can use the facet_wrap() function to divide the graphs in a single panel.
Adding the facet_wrap(~Nitrogen) code to the existing code will divide the graphs based on the variable Nitrogen.
ggplot (data=dataC, aes(x=variable, y=mean, fill=Nitrogen)) +
geom_bar(stat="identity",position="dodge") +
geom_errorbar(aes(ymin= mean- se, ymax=mean + se),
position=position_dodge(0.9), width=0.2) +
scale_fill_manual(values= c("Gray","Dark green")) +
scale_y_continuous(breaks= seq(0, 70, 10), limits= c(0, 70))+
facet_wrap(~Nitrogen) +
labs(fill="Nitrogen", x="Cultivar", y="Yield") +
theme(axis.title= element_text (face= "plain", size= 17, color= "black"),
axis.text.x= element_text(size= 15),
axis.text.y= element_text(size= 15),
axis.line= element_line(size= 0.5, colour= "black"),
legend.position= 'bottom',
legend.key= element_rect(color= "white", fill= "white"),
legend.key.size= unit(0.5,"cm"),
legend.title= element_text(face="plain", size= 14, color= "Black"),
legend.text= element_text(face="plain", size= 14, color= "Black"),
strip.text.x= element_text(size=17),
plot.margin=unit(c(0.5,0.5,0.5,0.5),"cm")) +
windows(width=7, height=5)
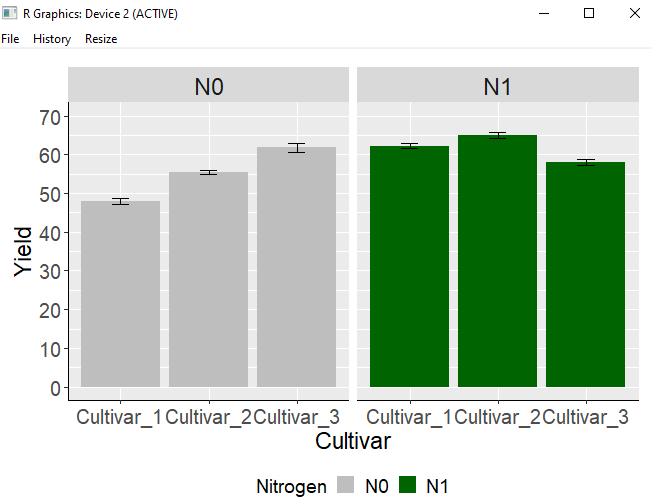
Then, you will be able to obtain a graph similar to the one described above.
Extra tips!!!
1) Adjusting the size of titles in panels
strip.text.x= element_text(size=17) By adding the above code to the complete set of code, you can modify the size of the label titles.
2) Changing the name of labels
newLable= c("Low Nitrogen", "High Nitrogen")
names(newLable)= c("N0", "N1")In this manner, a code is prepared beforehand to change N0 and N1 to “Low Nitrogen” and “High Nitrogen” before generating the ggplot, and Insert the labeller=labeller(Nitrogen=newLabel) code inside the facet_wrap() code.
you would write the code as follows:
newLable= c("Low Nitrogen", "High Nitrogen")
names(newLable)= c("N0", "N1")
##
ggplot (data=dataC, aes(x=variable, y=mean, fill=Nitrogen)) +
geom_bar(stat="identity",position="dodge") +
geom_errorbar(aes(ymin= mean- se, ymax=mean + se), position=position_dodge(0.9),
width=0.2) +
scale_fill_manual(values= c("Gray","Dark green")) +
scale_y_continuous(breaks= seq(0, 70, 10), limits= c(0, 70))+
facet_wrap(~Nitrogen, labeller=labeller(Nitrogen=newLable)) +
labs(fill="Nitrogen", x="Cultivar", y="Yield") +
theme(axis.title= element_text (face= "plain", size= 17, color= "black"),
axis.text.x= element_text(size= 15),
axis.text.y= element_text(size= 15),
axis.line= element_line(size= 0.5, colour= "black"),
legend.position= 'bottom',
legend.key= element_rect(color= "white", fill= "white"),
legend.key.size= unit(0.5,"cm"),
legend.title= element_text(face="plain", size= 14, color= "Black"),
legend.text= element_text(face="plain", size= 14, color= "Black"),
strip.text.x= element_text(size=25),
plot.margin=unit(c(0.5,0.5,0.5,0.5),"cm")) +
windows(width=7, height=5)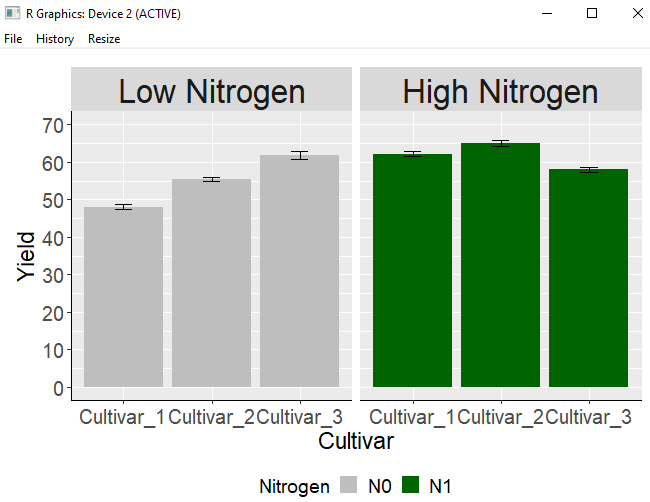
Another approach is to directly change the variable names in the data table. This is the method to modify the variable names within the Nitrogen column of the dataC dataset. I will change N0 to “0kg Nitrogen” and N1 to “200kg Nitrogen”.
Nitrogen variable mean sd n se
1 N0 Cultivar_1 48.0 1.581139 5 0.7071068
2 N0 Cultivar_2 55.4 1.140175 5 0.5099020
3 N0 Cultivar_3 61.8 2.387467 5 1.0677078
4 N1 Cultivar_1 62.2 1.483240 5 0.6633250
5 N1 Cultivar_2 65.0 1.581139 5 0.7071068
6 N1 Cultivar_3 58.0 1.581139 5 0.7071068
dataC$Nitrogen= factor(dataC$Nitrogen , levels = c("N0", "N1"),
labels= c("0kg Nitrogen", "200kg High Nitrogen"))Once the code runs, you will notice that the title names have been changed.
ggplot (data=dataC, aes(x=variable, y=mean, fill=Nitrogen)) +
geom_bar(stat="identity",position="dodge") +
geom_errorbar(aes(ymin= mean- se, ymax=mean + se), position=position_dodge(0.9),
width=0.2) +
scale_fill_manual(values= c("Gray","Dark green")) +
scale_y_continuous(breaks= seq(0, 70, 10), limits= c(0, 70))+
facet_wrap(~Nitrogen) +
labs(fill="Nitrogen", x="Cultivar", y="Yield") +
theme(axis.title= element_text (face= "plain", size= 17, color= "black"),
axis.text.x= element_text(size= 15),
axis.text.y= element_text(size= 15),
axis.line= element_line(size= 0.5, colour= "black"),
legend.position= 'bottom',
legend.key= element_rect(color= "white", fill= "white"),
legend.key.size= unit(0.5,"cm"),
legend.title= element_text(face="plain", size= 14, color= "Black"),
legend.text= element_text(face="plain", size= 14, color= "Black"),
strip.text.x= element_text(size=15),
plot.margin=unit(c(0.5,0.5,0.5,0.5),"cm")) +
windows(width=7, height=5)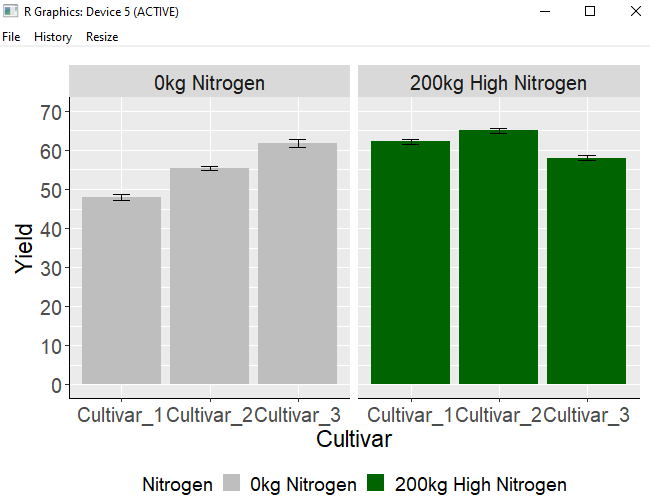
Another method is to use the stringr package to change the variable names within a column. If you execute the below code, you will be able to observe that the label names have been altered.
library (stringr)
dataC$Nitrogen= str_replace_all (dataC$Nitrogen, 'N0', 'No Fertilizer')
dataC$Nitrogen= str_replace_all (dataC$Nitrogen, 'N1', 'Fertilizer')3) Filling the label colors
strip.background= element_rect(color="Red", fill="Yellow", size=2, linetype="solid")Insert the above code. The label’s border color is red, the fill color is yellow, the border size is 2, and the border line is set to be solid.
ggplot (data=dataC, aes(x=variable, y=mean, fill=Nitrogen)) +
geom_bar(stat="identity",position="dodge") +
geom_errorbar(aes(ymin= mean- se, ymax=mean + se),
position=position_dodge(0.9), width=0.2) +
scale_fill_manual(values= c("Gray","Dark green")) +
scale_y_continuous(breaks= seq(0, 70, 10), limits= c(0, 70))+
facet_wrap(~Nitrogen) +
labs(fill="Nitrogen", x="Cultivar", y="Yield") +
theme(axis.title= element_text (face= "plain", size= 17, color= "black"),
axis.text.x= element_text(size= 15),
axis.text.y= element_text(size= 15),
axis.line= element_line(size= 0.5, colour= "black"),
legend.position= 'bottom',
legend.key= element_rect(color= "white", fill= "white"),
legend.key.size= unit(0.5,"cm"),
legend.title= element_text(face="plain", size= 14, color= "Black"),
legend.text= element_text(face="plain", size= 14, color= "Black"),
strip.text.x= element_text(size=15),
strip.background = element_rect(color="Red", fill="Yellow", size=2,
linetype="solid"),
plot.margin=unit(c(0.5,0.5,0.5,0.5),"cm")) +
windows(width=7, height=5)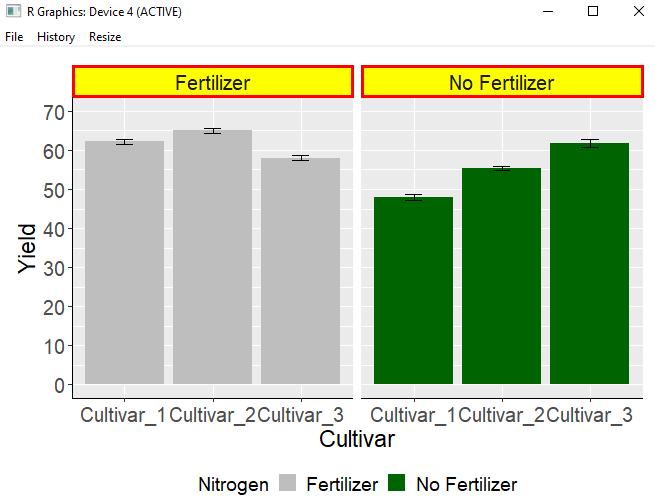
# full code
library (dplyr)
library (stringr)
library(ggplot2)
Nitrogen= c(rep("N0", 5), rep("N1", 5))
Cultivar_1= c(50,49,48,47,46,60,62,63,64,62)
Cultivar_2= c(55,57,56,55,54,65,66,67,64,63)
Cultivar_3= c(60,62,63,65,59,60,59,57,56,58)
dataA= data.frame(Nitrogen, Cultivar_1, Cultivar_2, Cultivar_3)
dataB= reshape2::melt(dataA[c("Nitrogen","Cultivar_1","Cultivar_2", "Cultivar_3")],
id.vars=c("Nitrogen"))
dataC= data.frame(dataB %>%
group_by(Nitrogen, variable) %>%
summarise(mean=mean(value), sd=sd(value), n=length(value), se=sd/sqrt(n)))
dataC$Nitrogen= str_replace_all (dataC$Nitrogen, 'N0', 'No Fertilizer')
dataC$Nitrogen= str_replace_all (dataC$Nitrogen, 'N1', 'Fertilizer')
ggplot (data=dataC, aes(x=variable, y=mean, fill=Nitrogen)) +
geom_bar(stat="identity",position="dodge") +
geom_errorbar(aes(ymin= mean- se, ymax=mean + se),
position=position_dodge(0.9), width=0.2) +
scale_fill_manual(values= c("Gray","Dark green")) +
scale_y_continuous(breaks= seq(0, 70, 10), limits= c(0, 70))+
facet_wrap(~Nitrogen) +
labs(fill="Nitrogen", x="Cultivar", y="Yield") +
theme(axis.title= element_text (face= "plain", size= 17, color= "black"),
axis.text.x= element_text(size= 15),
axis.text.y= element_text(size= 15),
axis.line= element_line(size= 0.5, colour= "black"),
legend.position= 'bottom',
legend.key= element_rect(color= "white", fill= "white"),
legend.key.size= unit(0.5,"cm"),
legend.title= element_text(face="plain", size= 14, color= "Black"),
legend.text= element_text(face="plain", size= 14, color= "Black"),
strip.text.x= element_text(size=15),
strip.background = element_rect(color="Red", fill="Yellow", size=2,
linetype="solid"),
plot.margin=unit(c(0.5,0.5,0.5,0.5),"cm")) +
windows(width=7, height=5)<full code access available> https://github.com/agronomy4future/r_code/blob/main/facet_wrap
