Machine Learning: Predicting Values with Multiple Models- Part II
In my previous post, I predicted grain weight from length and width of grains using Random Forest.
■ Machine Learning: Predicting Values with Multiple Models- Part I
Now, my next question is how the model accuracy changes when grain area and genotype are added. If you followed my previous post closely, you should be able to understand the code below.
■ Data upload
import pandas as pd
import requests
from io import StringIO
github="https://raw.githubusercontent.com/agronomy4future/raw_data_practice/main/ML_Practice_Grain_Dimension.csv"
response=requests.get(github)
df=pd.read_csv(StringIO(response.text))df['Genotype'] = df['Genotype'].replace({'cv1': 0, 'cv2': 1, 'cv3': 2, 'cv4': 3, 'cv5': 4})
print(df.head(5))
Genotype Length_mm Width_mm Area_mm_2 GW_mg
0 0 4.86 2.12 7.56 14.44
1 0 5.01 1.94 7.25 13.67
2 0 5.54 1.97 7.99 15.55
3 0 3.84 1.99 5.86 10.32
4 0 5.30 2.06 8.04 15.66■ Data Splitting
# input
input= df.drop(columns=['GW_mg'])
print(input.head(5))
Genotype Length_mm Width_mm Area_mm_2
0 0 4.86 2.12 7.56
1 0 5.01 1.94 7.25
2 0 5.54 1.97 7.99
3 0 3.84 1.99 5.86
4 0 5.30 2.06 8.04
# target
target= df['GW_mg']
print(target.head(5))
0 14.44
1 13.67
2 15.55
3 10.32
4 15.66Unlike the previous data, I have now added genotype and grain area to the model.
# Splitting the dataset into training and testing sets
from sklearn.model_selection import train_test_split
train_input, test_input, train_target, test_target = train_test_split(input, target, test_size=0.3, random_state=42)■ Machine Learning Algorithm
1. Random Forest
from sklearn.ensemble import RandomForestRegressor
rf= RandomForestRegressor()
rf_model= rf.fit(train_input, train_target)
accuracy= rf_model.score(test_input, test_target)
print("Accuracy:", accuracy)
Accuracy: 0.99999918922536992. K-nearest neighbors
from sklearn.neighbors import KNeighborsRegressor
knr = KNeighborsRegressor()
knr_model = knr.fit(train_input, train_target)
accuracy = knr_model.score(test_input, test_target)
print("Accuracy:", accuracy)
Accuracy: 0.99993468027432533. Linear Regression
from sklearn.linear_model import LinearRegression
lr= LinearRegression ()
lr_model = lr.fit(train_input, train_target)
accuracy = lr_model.score(test_input, test_target)
print("Accuracy:", accuracy)
Accuracy: 0.99777571358496834. Decision Tree
from sklearn.tree import DecisionTreeRegressor
dt = DecisionTreeRegressor()
dt_model= dt.fit(train_input, train_target)
accuracy = dt_model.score(test_input, test_target)
print("Accuracy:", accuracy)
Accuracy: 0.99999854753342755. Ridge regression
from sklearn.linear_model import Ridge
ridge = Ridge()
ridge_model= ridge.fit(train_input, train_target)
accuracy = ridge_model.score(test_input, test_target)
print("Accuracy:", accuracy)
Accuracy: 0.9977757295600886. Lasso regression
from sklearn.linear_model import Lasso
lasso = Lasso()
lasso_model= lasso.fit(train_input, train_target)
accuracy = lasso_model.score(test_input, test_target)
print("Accuracy:", accuracy)
Accuracy: 0.9969952312548194Let’s compare the accuracy when using length and width data versus when using length, width, area, and genotype data. The accuracy increased overall when using length, width, area, and genotype data. I’ll choose the Random Forest model again.
| Model | Accuracy (using length, width) | Accuracy (using length, width, area and genotype) |
| Random Forest | 0.967 | 0.999 |
| K-nearest neighbors | 0.963 | 0.999 |
| Linear Regression | 0.878 | 0.998 |
| Decision Tree | 0.955 | 0.999 |
| Ridge | 0.878 | 0.998 |
| Lasso | 0.848 | 0.997 |
■ To verify the model
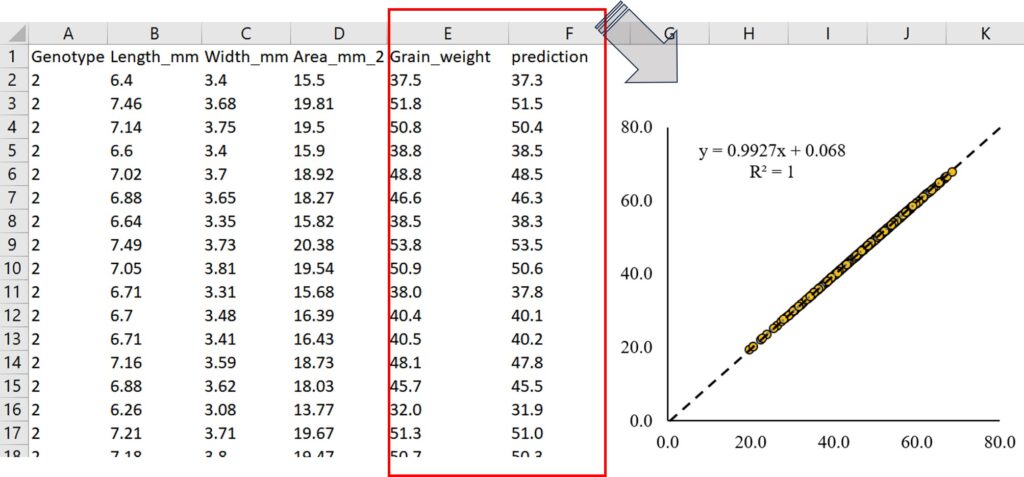
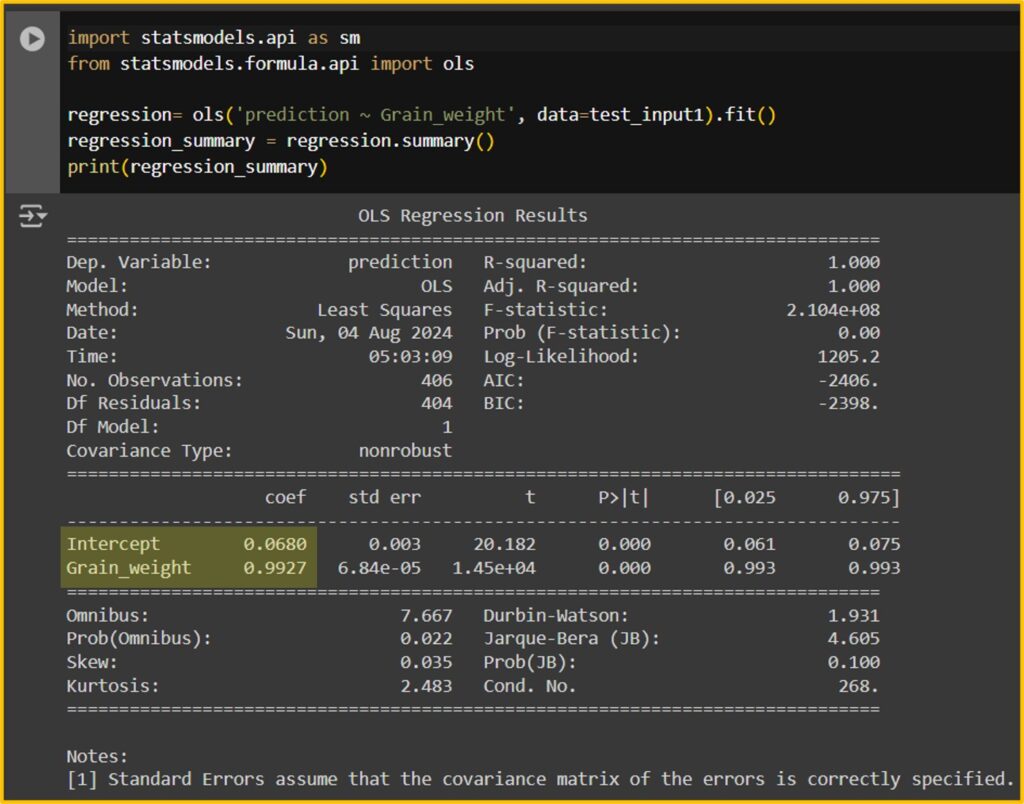
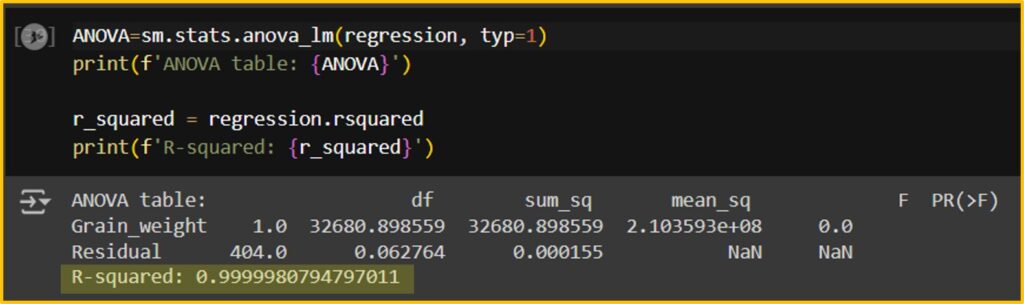
Now, I’ll use a new dataset to compare the actual grain weight with the grain weight predicted by the random forest model.
github="https://raw.githubusercontent.com/agronomy4future/raw_data_practice/main/ML_Practice_Grain_Dimension_Verifying.csv"
response=requests.get(github)
df1=pd.read_csv(StringIO(response.text))
print(df1.head(5))
Genotype Length_mm Width_mm Area_mm_2 GW_mg
0 cv3 6.40 3.40 15.50 37.46
1 cv3 7.46 3.68 19.81 51.82
2 cv3 7.14 3.75 19.50 50.76
3 cv3 6.60 3.40 15.90 38.75
4 cv3 7.02 3.70 18.92 48.77df1['Genotype'] = df1['Genotype'].replace({'cv3': 2, 'cv4': 3, 'cv5': 4})
print(df1.head(5))
Genotype Length_mm Width_mm Area_mm_2 GW_mg
0 2 6.40 3.40 15.50 37.46
1 2 7.46 3.68 19.81 51.82
2 2 7.14 3.75 19.50 50.76
3 2 6.60 3.40 15.90 38.75
4 2 7.02 3.70 18.92 48.77Next, I’ll split the data into training and testing sets using this new dataset.
test_input1= df1.drop(columns=['GW_mg'])
print(test_input1.head(5))
Genotype Length_mm Width_mm Area_mm_2
0 2 6.40 3.40 15.50
1 2 7.46 3.68 19.81
2 2 7.14 3.75 19.50
3 2 6.60 3.40 15.90
4 2 7.02 3.70 18.92GW_mg = df1['GW_mg']
test_target1 = GW_mg
print(test_target1.head(5))
0 37.46
1 51.82
2 50.76
3 38.75
4 48.77Random Forest
from sklearn.ensemble import RandomForestRegressor
rf= RandomForestRegressor(random_state=42)
rf_model= rf.fit(train_input, train_target)
accuracy= rf_model.score(test_input1, test_target1)
print("Accuracy:", accuracy)
Accuracy: 0.9989566381418276I’ll add predicted grain weight data.
y_pred= lasso_model.predict(test_input1)
test_input1['prediction']= y_pred
print(test_input1.head(5))
Genotype Length_mm Width_mm Area_mm_2 prediction
0 2 6.40 3.40 15.50 37.269362
1 2 7.46 3.68 19.81 51.501729
2 2 7.14 3.75 19.50 50.442392
3 2 6.60 3.40 15.90 38.539158
4 2 7.02 3.70 18.92 48.479086test_input1['Grain_weight']= test_target1
print(test_input1.head(5))
Genotype Length_mm Width_mm Area_mm_2 prediction Grain_weight
0 2 6.40 3.40 15.50 37.269362 37.46
1 2 7.46 3.68 19.81 51.501729 51.82
2 2 7.14 3.75 19.50 50.442392 50.76
3 2 6.60 3.40 15.90 38.539158 38.75
4 2 7.02 3.70 18.92 48.479086 48.77Now, actual and predicted grain weight are added in test_input1 data. I’ll download this data to my PC.
import pandas as pd
from google.colab import files
# Save the DataFrame to a CSV file
test_input1.to_csv('Grain_weight_2024.csv', index=False)
# Use the files module to download the file
files.download('Grain_weight_2024.csv')Now the predicted grain weight is highly accurate.