Converting Rows to Columns in R: A Guide to Transposing Data (feat. pivot_wider and pivot_longer)
When data is arranged, it can be structured either vertically (row-based) or horizontally (column-based). The choice depends on your preference for organizing data. However, when running statistics, data should be arranged row-based, as variables need to be in the same column. On the other hand, when calculating per variable, it is much easier to organize data column-based, allowing for simpler calculations. Regardless of the approach, well-organized data is essential, and the ability to restructure data is a valuable skill. Today, I will introduce how to automatically reshape data both vertically and horizontally using R.
Let’s upload a dataset in R.
library(readr)
github="https://raw.githubusercontent.com/agronomy4future/raw_data_practice/main/wheat_grain_Fe_uptake.csv"
dataA= data.frame(read_csv(url(github), show_col_types= FALSE))
Location Season Genotype Reps Iron_ton_ha Stage Fe
1 East 2021 CV1 1 21.7127 Vegetative 0.44
2 East 2021 CV1 2 8.7340 Vegetative 0.30
3 East 2021 CV1 3 9.5003 Vegetative 0.31
4 East 2021 CV1 4 5.9481 Vegetative 0.37
5 East 2021 CV1 5 7.4608 Vegetative 0.30
6 East 2021 CV1 6 20.5326 Vegetative 0.33
7 East 2021 CV1 7 19.8532 Vegetative 0.29
8 East 2021 CV1 8 7.9718 Vegetative 0.35
9 East 2021 CV1 9 15.0087 Vegetative 0.38
10 East 2021 CV1 10 5.6608 Vegetative 0.40
.
.
.Now, I’d like to move the variables in the ‘Stage’ column to individual columns. I’ll use pivot_wider()
library(dplyr)
library(tidyr)
dataB= data.frame(dataA %>%
group_by(Location, Season, Genotype, Iron_ton_ha, Stage) %>%
pivot_wider(names_from= Stage, values_from=Fe))
dataB
Location Season Genotype Reps Iron_ton_ha Vegetative Reproductive Maturity
1 East 2021 CV1 1 21.7127 0.44 0.37 0.13
2 East 2021 CV1 2 8.7340 0.30 0.32 0.14
3 East 2021 CV1 3 9.5003 0.31 0.33 0.12
4 East 2021 CV1 4 5.9481 0.37 0.30 0.18
5 East 2021 CV1 5 7.4608 0.30 0.34 0.12
6 East 2021 CV1 6 20.5326 0.33 0.33 0.24
7 East 2021 CV1 7 19.8532 0.29 0.31 0.21
8 East 2021 CV1 8 7.9718 0.35 0.31 0.17
9 East 2021 CV1 9 15.0087 0.38 0.30 0.32
10 East 2021 CV1 10 5.6608 0.40 0.31 0.14Data was re-structured as horizontally. Let’s change it as vertically again.
dataC= data.frame(dataB %>%
pivot_longer(
cols= c(Vegetative, Reproductive, Maturity),
names_to= "Stage",
values_to= "Fe"))
dataC
Location Season Genotype Reps Iron_ton_ha Stage Fe
1 East 2021 CV1 1 21.7127 Vegetative 0.44
2 East 2021 CV1 1 21.7127 Reproductive 0.37
3 East 2021 CV1 1 21.7127 Maturity 0.13
4 East 2021 CV1 2 8.7340 Vegetative 0.30
5 East 2021 CV1 2 8.7340 Reproductive 0.32
6 East 2021 CV1 2 8.7340 Maturity 0.14
7 East 2021 CV1 3 9.5003 Vegetative 0.31
8 East 2021 CV1 3 9.5003 Reproductive 0.33
9 East 2021 CV1 3 9.5003 Maturity 0.12
10 East 2021 CV1 4 5.9481 Vegetative 0.37
.
.
.Practice
Here is a practice for you.
library(readr)
github="https://raw.githubusercontent.com/agronomy4future/raw_data_practice/main/yield_per_location.csv"
dataA=data.frame(read_csv(url(github), show_col_types=FALSE))
dataA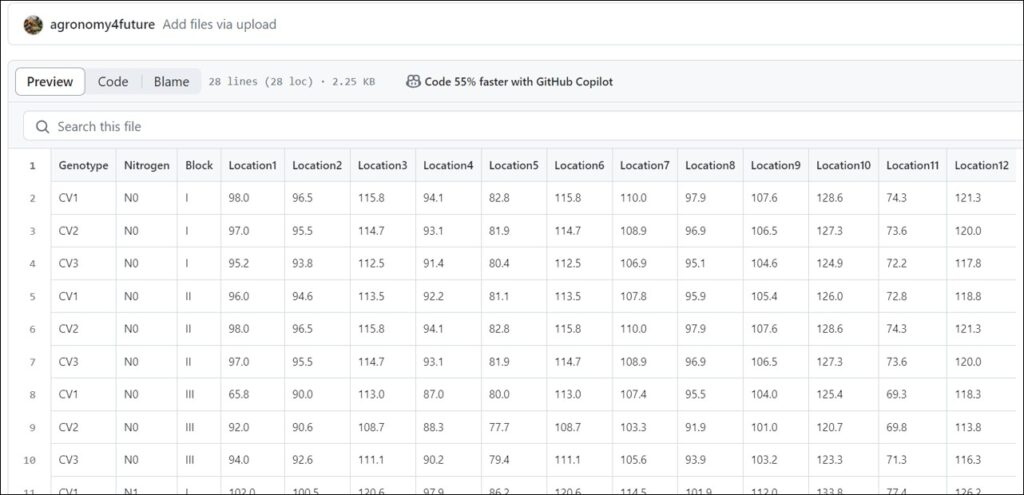
You can transpose the columns to rows using pivot_longer(). The answer code is in the below code sumamry.
full code: https://github.com/agronomy4future/r_code/blob/main/Converting_Rows_to_Columns_in_R_A_Guide_to_Transposing_Data_(feat_pivot_wider_and_pivot_longer).ipynb
